Identification of Salvianolic acid A as a potent inhibitor of PDEs to enhance proliferation of human neural stem cells
IF 4
2区 化学
Q2 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
The inhibition of phosphodiesterases (PDEs) is a promising therapeutic strategy for treating central nervous system (CNS) disorders due to its capacity to facilitate neuroplasticity. Salvianolic acid A (SAA) shows promise in treating CNS disorders, but its specific targets are still unclear. This study has discovered that SAA directly targets PDEs to promote the proliferation of neural stem cells (NSCs). A pharmacophore model for PDE9 inhibitors was developed and validated to screen a compound database, leading to the discovery of SAA as an active agent. Further investigation into the anti-PDEs activity of SAA has revealed that it is a broad-spectrum PDEs inhibitor, with IC50 values of 28.26, 34.18, and 31.68 µM for PDE4, PDE5, and PDE9, respectively. Furthermore, the proliferation of human NSCs (H9) was significantly enhanced by 30–90 µM SAA. This study also provides comprehensive elucidations of signaling pathways associated with direct targeting of PDEs for neuroplasticity in stroke. Furthermore, molecular dynamics (MD) simulations revealed that SAA effectively interacts with the crucial amino acid residues of PDE9. The decomposition of binding free energy for acid residues offers further insight into the interactions between SAA and these key amino acids, thus providing more information for understanding the mechanism of SAA bound to PDE9.
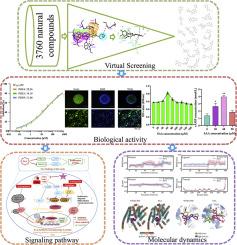
丹酚酸A作为PDEs有效抑制剂促进人神经干细胞增殖的鉴定
抑制磷酸二酯酶(PDEs)因其促进神经可塑性的能力而成为治疗中枢神经系统(CNS)疾病的一种有前景的治疗策略。丹酚酸A (SAA)有望治疗中枢神经系统疾病,但其具体靶点尚不清楚。本研究发现SAA可直接作用于PDEs,促进神经干细胞(NSCs)的增殖。开发了PDE9抑制剂的药效团模型,并对其进行了验证,以筛选化合物数据库,从而发现SAA是一种活性剂。对SAA抗PDEs活性的进一步研究表明,SAA是一种广谱PDEs抑制剂,对PDE4、PDE5和PDE9的IC50值分别为28.26、34.18和31.68µM。此外,30-90µM SAA能显著促进人NSCs (H9)的增殖。该研究还提供了与PDEs直接靶向脑卒中神经可塑性相关的信号通路的全面阐明。此外,分子动力学(MD)模拟表明,SAA有效地与PDE9的关键氨基酸残基相互作用。酸残基结合自由能的分解有助于进一步了解SAA与这些关键氨基酸的相互作用,从而为了解SAA与PDE9结合的机制提供更多信息。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Journal of Molecular Structure
化学-物理化学
CiteScore
7.10
自引率
15.80%
发文量
2384
审稿时长
45 days
期刊介绍:
The Journal of Molecular Structure is dedicated to the publication of full-length articles and review papers, providing important new structural information on all types of chemical species including:
• Stable and unstable molecules in all types of environments (vapour, molecular beam, liquid, solution, liquid crystal, solid state, matrix-isolated, surface-absorbed etc.)
• Chemical intermediates
• Molecules in excited states
• Biological molecules
• Polymers.
The methods used may include any combination of spectroscopic and non-spectroscopic techniques, for example:
• Infrared spectroscopy (mid, far, near)
• Raman spectroscopy and non-linear Raman methods (CARS, etc.)
• Electronic absorption spectroscopy
• Optical rotatory dispersion and circular dichroism
• Fluorescence and phosphorescence techniques
• Electron spectroscopies (PES, XPS), EXAFS, etc.
• Microwave spectroscopy
• Electron diffraction
• NMR and ESR spectroscopies
• Mössbauer spectroscopy
• X-ray crystallography
• Charge Density Analyses
• Computational Studies (supplementing experimental methods)
We encourage publications combining theoretical and experimental approaches. The structural insights gained by the studies should be correlated with the properties, activity and/ or reactivity of the molecule under investigation and the relevance of this molecule and its implications should be discussed.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


