CLMS: Bridging domain gaps in medical imaging segmentation with source-free continual learning for robust knowledge transfer and adaptation
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
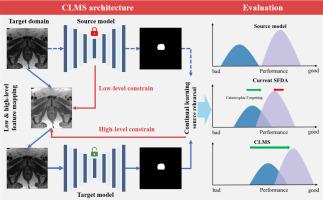
Deep learning shows promise for medical image segmentation but suffers performance declines when applied to diverse healthcare sites due to data discrepancies among the different sites. Translating deep learning models to new clinical environments is challenging, especially when the original source data used for training is unavailable due to privacy restrictions. Source-free domain adaptation (SFDA) aims to adapt models to new unlabeled target domains without requiring access to the original source data. However, existing SFDA methods face challenges such as error propagation, misalignment of visual and structural features, and inability to preserve source knowledge. This paper introduces Continual Learning Multi-Scale domain adaptation (CLMS), an end-to-end SFDA framework integrating multi-scale reconstruction, continual learning, and style alignment to bridge domain gaps across medical sites using only unlabeled target data or publicly available data. Compared to the current state-of-the-art methods, CLMS consistently and significantly achieved top performance for different tasks, including prostate MRI segmentation (improved Dice of 10.87 %), colonoscopy polyp segmentation (improved Dice of 17.73 %), and plus disease classification from retinal images (improved AUC of 11.19 %). Crucially, CLMS preserved source knowledge for all the tasks, avoiding catastrophic forgetting. CLMS demonstrates a promising solution for translating deep learning models to new clinical imaging domains towards safe, reliable deployment across diverse healthcare settings.
CLMS:通过无源持续学习弥合医学成像分割领域的差距,实现强大的知识转移和适应
深度学习显示了医学图像分割的前景,但由于不同站点之间的数据差异,当应用于不同的医疗保健站点时,性能会下降。将深度学习模型转化为新的临床环境是具有挑战性的,特别是当用于训练的原始源数据由于隐私限制而不可用时。无源域自适应(SFDA)旨在使模型适应新的未标记的目标域,而不需要访问原始源数据。然而,现有的SFDA方法面临着诸如误差传播、视觉和结构特征不一致以及无法保存源知识等挑战。本文介绍了持续学习多尺度域适应(CLMS),这是一个端到端的SFDA框架,集成了多尺度重建、持续学习和风格对齐,仅使用未标记的目标数据或公开可用的数据来弥合医疗站点之间的域差距。与目前最先进的方法相比,CLMS在不同任务上一致且显著地取得了最佳性能,包括前列腺MRI分割(提高了10.87%的Dice),结肠镜息肉分割(提高了17.73%的Dice),以及视网膜图像的疾病分类(提高了11.19%的AUC)。关键是,CLMS保留了所有任务的源知识,避免了灾难性遗忘。CLMS展示了一种很有前途的解决方案,可将深度学习模型转换为新的临床成像领域,从而在各种医疗保健环境中安全、可靠地部署。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


