ROC-guided virtual screening, molecular dynamics simulation, and bioactivity validation assessment Z195914464 as a 3CL Mpro inhibitor
IF 2.2
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Discovering novel class anti-SARS-CoV-2 compounds with novel backbones is essential for preventing and controlling SARS-CoV-2 transmission, which poses a substantial threat to the health and social sustainable development of the global population because of its high pathogenicity and high transmissibility. Although the potential mutation of SARS-CoV-2 might diminish the therapeutic efficacy of drugs, 3CL Mpro is the target highly conservative in contrast with other targets. It is an essential enzyme for coronavirus replication. Based on this, this study utilized the drug discovery strategy of Knime molecular filtering framework, ROC-guided virtual screening, clustering analysis, binding mode analysis, and activity evaluation approaches to identify compound Z195914464 (IC50: 7.19 μM) is a novel class inhibitor of anti-SARS-CoV-2 against the 3CL Mpro target. In addition, based on molecular dynamics simulations and MMPBSA analyses, discovered that compound Z195914464 can interact with more key residues and lower bonding energies, which explains why it exhibited more activity than the other three compounds. In summary, this study developed a method for the rapid and accurate discovery of active compounds and can also be applied in the discovery of active compounds in other targets.
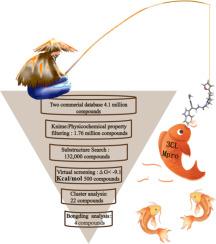
Z195914464作为3CL Mpro抑制剂的虚拟筛选、分子动力学模拟和生物活性验证评估
SARS-CoV-2具有高致病性和高传播性,对全球人口的健康和社会可持续发展构成重大威胁,发现具有新型骨架的新型抗SARS-CoV-2化合物是预防和控制SARS-CoV-2传播的关键。尽管SARS-CoV-2的潜在突变可能会降低药物的治疗效果,但与其他靶点相比,3CL Mpro是高度保守的靶点。它是冠状病毒复制的必需酶。基于此,本研究利用Knime分子过滤框架的药物发现策略、roc引导虚拟筛选、聚类分析、结合模式分析和活性评价等方法,鉴定化合物Z195914464 (IC50: 7.19 μM)是一种针对3CL Mpro靶点的新型抗sars - cov -2类抑制剂。此外,基于分子动力学模拟和MMPBSA分析,发现化合物Z195914464可以与更多的关键残基相互作用,键能较低,这解释了为什么它比其他三个化合物表现出更强的活性。综上所述,本研究开发了一种快速准确发现活性化合物的方法,也可应用于其他靶标中活性化合物的发现。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Biophysical chemistry
生物-生化与分子生物学
CiteScore
6.10
自引率
10.50%
发文量
121
审稿时长
20 days
期刊介绍:
Biophysical Chemistry publishes original work and reviews in the areas of chemistry and physics directly impacting biological phenomena. Quantitative analysis of the properties of biological macromolecules, biologically active molecules, macromolecular assemblies and cell components in terms of kinetics, thermodynamics, spatio-temporal organization, NMR and X-ray structural biology, as well as single-molecule detection represent a major focus of the journal. Theoretical and computational treatments of biomacromolecular systems, macromolecular interactions, regulatory control and systems biology are also of interest to the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


