Keep it accurate and robust: An enhanced nuclei analysis framework
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
Computational and structural biotechnology journal
Pub Date : 2024-11-13
DOI:10.1016/j.csbj.2024.10.046
引用次数: 0
Abstract
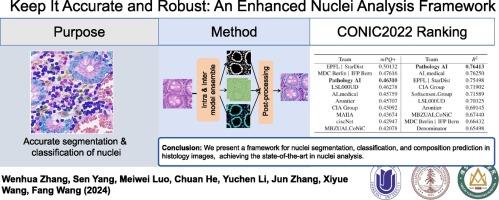
Accurate segmentation and classification of nuclei in histology images is critical but challenging due to nuclei heterogeneity, staining variations, and tissue complexity. Existing methods often struggle with limited dataset variability, with patches extracted from similar whole slide images (WSI), making models prone to falling into local optima. Here we propose a new framework to address this limitation and enable robust nuclear analysis. Our method leverages dual-level ensemble modeling to overcome issues stemming from limited dataset variation. Intra-ensembling applies diverse transformations to individual samples, while inter-ensembling combines networks of different scales. We also introduce enhancements to the HoVer-Net architecture, including updated encoders, nested dense decoding and model regularization strategy. We achieve state-of-the-art results on public benchmarks, including 1st place for nuclear composition prediction and 3rd place for segmentation/classification in the 2022 Colon Nuclei Identification and Counting (CoNIC) Challenge. This success validates our approach for accurate histological nuclei analysis. Extensive experiments and ablation studies provide insights into optimal network design choices and training techniques. In conclusion, this work proposes an improved framework advancing the state-of-the-art in nuclei analysis. We will release our code and models to serve as a toolkit for the community.
保持准确性和稳健性增强型原子核分析框架
对组织学图像中的细胞核进行精确分割和分类至关重要,但由于细胞核的异质性、染色变化和组织复杂性,这一工作极具挑战性。现有的方法往往难以应对有限的数据集变化,从相似的整张切片图像(WSI)中提取的斑块,使模型容易陷入局部最优。在此,我们提出了一个新的框架来解决这一局限性,并实现稳健的核分析。我们的方法利用双层集合建模来克服因数据集变化有限而产生的问题。集合内对单个样本进行不同的转换,而集合间则结合不同规模的网络。我们还对 HoVer-Net 架构进行了改进,包括更新编码器、嵌套密集解码和模型正则化策略。我们在公开基准测试中取得了最先进的结果,包括在 2022 年结肠核识别与计数(CoNIC)挑战赛中获得核成分预测第一名和分割/分类第三名。这一成功验证了我们准确分析组织学细胞核的方法。广泛的实验和消融研究为最佳网络设计选择和训练技术提供了启示。总之,这项工作提出了一个改进的框架,推动了细胞核分析技术的发展。我们将发布我们的代码和模型,作为社区的工具包。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational and structural biotechnology journal
Biochemistry, Genetics and Molecular Biology-Biophysics
CiteScore
9.30
自引率
3.30%
发文量
540
审稿时长
6 weeks
期刊介绍:
Computational and Structural Biotechnology Journal (CSBJ) is an online gold open access journal publishing research articles and reviews after full peer review. All articles are published, without barriers to access, immediately upon acceptance. The journal places a strong emphasis on functional and mechanistic understanding of how molecular components in a biological process work together through the application of computational methods. Structural data may provide such insights, but they are not a pre-requisite for publication in the journal. Specific areas of interest include, but are not limited to:
Structure and function of proteins, nucleic acids and other macromolecules
Structure and function of multi-component complexes
Protein folding, processing and degradation
Enzymology
Computational and structural studies of plant systems
Microbial Informatics
Genomics
Proteomics
Metabolomics
Algorithms and Hypothesis in Bioinformatics
Mathematical and Theoretical Biology
Computational Chemistry and Drug Discovery
Microscopy and Molecular Imaging
Nanotechnology
Systems and Synthetic Biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


