Discrete element method-based simulation system for predicting natural stone evacuation pathways in patients with urolithiasis
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
Abstract
Urolithiasis is a globally prevalent disease with high incidence and recurrence rates and is often accompanied by severe pain. Its ideal treatment is spontaneous stone passage, avoiding the invasiveness associated with surgery. However, the mechanisms underlying spontaneous stone passage remain unclear. Therefore, in this study, we developed a kidney stone trajectory prediction simulation system using the discrete element method (DEM) to elucidate spontaneous passage mechanisms by analyzing and visualizing stone trajectories within the kidney. We compared this simulation system with experiments using a three-dimensional kidney replica of patients with urolithiasis to optimize critical DEM parameters, including the collision margin ε, friction coefficient Cf, and restitution coefficient Cr. The reliability of these optimized parameters was validated using kidney shapes that differed from those used in the optimization experiments. The simulation system with optimized parameters consistently demonstrated high fidelity to the experimental results, regardless of kidney shape, initial stone position, or stone size. These findings demonstrate the reliability of the simulation system, underscoring its potential contribution to developing new and effective treatments for urolithiasis by improving the accuracy of stone trajectory predictions.
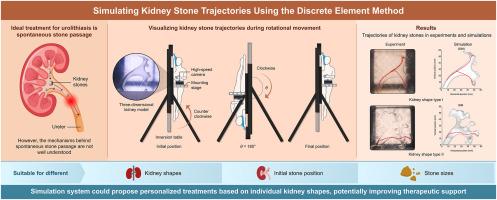
基于离散元方法的模拟系统,用于预测泌尿系统结石患者体内天然结石的排空路径
尿路结石是一种全球流行的疾病,发病率和复发率都很高,而且常常伴有剧烈疼痛。理想的治疗方法是让结石自发排出,避免手术带来的创伤。然而,自发排石的机制仍不清楚。因此,在本研究中,我们利用离散元法(DEM)开发了一个肾结石轨迹预测模拟系统,通过分析和可视化结石在肾脏内的轨迹来阐明自发性排石机制。我们将该模拟系统与使用尿路结石患者三维肾脏复制品进行的实验进行了比较,以优化关键的 DEM 参数,包括碰撞裕度 ε、摩擦系数 Cf 和复原系数 Cr。使用与优化实验中不同的肾脏形状验证了这些优化参数的可靠性。无论肾脏形状、结石初始位置或结石大小如何,采用优化参数的模拟系统始终表现出与实验结果的高度保真。这些研究结果证明了模拟系统的可靠性,强调了它通过提高结石轨迹预测的准确性,为开发新的、有效的尿路结石治疗方法做出的潜在贡献。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


