Similarities between Ixodes ricinus and Ixodes inopinatus genomes and horizontal gene transfer from their endosymbionts
IF 1.7
Q3 PARASITOLOGY
Current research in parasitology & vector-borne diseases
Pub Date : 2024-01-01
DOI:10.1016/j.crpvbd.2024.100229
引用次数: 0
Abstract
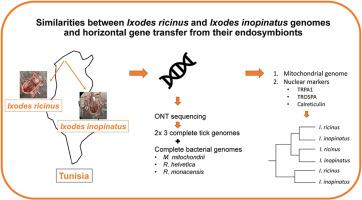
The taxa Ixodes ricinus and Ixodes inopinatus are sympatric in Tunisia. The genetics underlying their morphological differences are unresolved. In this study, ticks collected in Jouza-Amdoun, Tunisia, were morphologically identified and sequenced using Oxford Nanopore Technologies. Three complete genome assemblies of I. inopinatus and three of I. ricinus with BUSCO scores of ∼98% were generated, including the reconstruction of mitochondrial genomes and separation of both alleles of the TRPA1, TROSPA and calreticulin genes. Deep sequencing allowed the first descriptions of complete bacterial genomes for “Candidatus Midichloria mitochondrii”, Rickettsia helvetica and R. monacensis from North Africa, and the discovery of extensive integration of parts of the Spiroplasma ixodetis and “Ca. M. mitochondrii” into the nuclear genome of these ticks. Phylogenetic analyses of the mitochondrial genome, the nuclear genes, and symbionts showed differentiation between Tunisian and Dutch ticks, but high genetic similarities between Tunisian I. ricinus and I. inopinatus. Subtraction of the genome assemblies identified the presence of some unique sequences, which could not be confirmed when screening a larger batch of I. ricinus and I. inopinatus ticks using PCR. Our findings yield compelling evidence that I. inopinatus is genetically highly similar, if not identical, to sympatric I. ricinus. Defined morphological differences might be caused by extrinsic factors such as micro-climatic conditions or bloodmeal composition. Our findings support the existence of different lineages of I. ricinus as well of its symbionts/pathogens from geographically dispersed locations.
Ixodes ricinus 和 Ixodes inopinatus 基因组的相似性及其内共生体的水平基因转移
Ixodes ricinus 和 Ixodes inopinatus 是突尼斯的同域类群。它们形态差异的遗传学基础尚未解决。在这项研究中,对在突尼斯 Jouza-Amdoun 采集的蜱虫进行了形态鉴定,并使用牛津纳米孔技术进行了测序。结果显示,I. inopinatus 和 I. ricinus 的三个基因组组装完整,BUSCO 得分为 98%,其中包括线粒体基因组的重建以及 TRPA1、TROSPA 和 calreticulin 基因两个等位基因的分离。深度测序首次描述了北非 "线粒体 Midichloria 样菌"、Rickettsia helvetica 和 R. monacensis 的完整细菌基因组,并发现了 Spiroplasma ixodetis 和 "Ca. M. mitochondrii "的部分基因广泛整合。在这些蜱虫的核基因组中发现了线粒体螺旋体和 "Ca. M. mitochondrii "的广泛整合。线粒体基因组、核基因和共生体的系统发育分析表明,突尼斯蜱和荷兰蜱之间存在差异,但突尼斯蓖麻蜱和inopinatus蜱之间的遗传相似性很高。基因组组装的减法发现了一些独特的序列,但在使用 PCR 对更大一批 I. ricinus 和 I. inopinatus 蜱进行筛选时,这些序列无法得到证实。我们的研究结果提供了令人信服的证据,表明 I. inopinatus 与同域 I. ricinus 在基因上高度相似,甚至完全相同。明确的形态差异可能是由微气候条件或血餐成分等外在因素造成的。我们的研究结果表明,蓖麻蛙存在不同的品系,其共生体/病原体也来自地理上分散的地点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


