Automatic discrimination between neuroendocrine carcinomas and grade 3 neuroendocrine tumors by deep learning of H&E images
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
Abstract
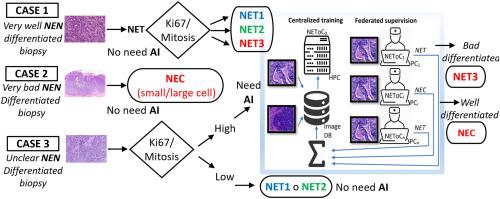
Neuroendocrine neoplasms (NENs) arise from diffuse neuroendocrine cells and are categorized as either well-differentiated and less proliferative Neuroendocrine Tumors (NETs), divided into low (G1), middle (G2), and high grades (G3), or poorly differentiated, and more proliferative Neuroendocrine Carcinomas (NECs). Low-grade NENs typically necessitate surgical intervention, whereas high-grade ones often require chemotherapy. However, low-grade NENs may exhibit aggressive behavior. Therefore, it is crucial to precisely refine the diagnosis of NENs. This refinement is achievable when differentiation/non-differentiation is evident or when the Ki-67 or mitosis index is low. The challenge arises in cases of morphologically undifferentiated instances with a high Ki-67 percentage and/or high mitotic index. To address this challenge, we developed a Deep Learning (DL) system named NEToC, designed to differentiate between NETs and NECs using exclusively morphological information from immunohistochemistry images, without relying on Ki-67 or mitosis assessments. NEToC was developed using 95 NEN cases from the period 2015 to 2018 at Parc Tauli Hospital in Spain, comprising 588 images. Implemented as a Graphical User Interface (GUI) system, NEToC is intended for deployment in pathological departments of hospitals to perform federated supervision. We tested the performance of NEToC with 119 images that were not used during the Artificial Neural Network (ANN) training phase, and evaluated its robustness across various resolutions: 64 × 64, 128 × 128, 256 × 256, and 512 × 512 pixels. The achieved accuracies for these resolutions were 74 %, 98 %, 98 %, and 100 %, respectively, for an underrepresented NET G3 experiment, and 66 %, 89 %, 95 % and 94 % for a represented NET G3 experiment. Based on several measured performance metrics, the optimal resolution appears to be between 128 × 128 and 256 × 256 pixels, considering computational resources and accuracy requirements. However, we found that the 256 × 256-pixel resolution is more robust to classify underrepresented classes in the learning phase. These results imply that the information to discriminate between NECs and Grade 3 NETs needs to be resolved in regions with a pixel resolution of no more than 4 μm/pixel. Most of the misclassifications were false negatives, where NET G1-type images were erroneously classified as NEC-type. Our results demonstrate that a DL-based diagnostic algorithm provides a more accurate diagnosis in NEN cases where physicians face challenges. NEToC has been initially trained with and used to classify gastrointestinal NENs. Since the NEN morphology does not change among the different organs, the use of NEToC can be extrapolated to NENs from different organs. NEToC facilitates federated supervision, allowing pathologists to collect interchangeable files based on NEToC classification predictions. NEToC is an easy-to-use, adaptable software that integrates multiple ANNs to improve standardization and accuracy NEN diagnosis, opening up possibilities for combining DL and histological diagnosis in federated supervision systems. A future goal is to classify not only NETs, but also the three-tier system (NET G1, NET G2, and NET G3) based solely on tissue differentiation information.
通过对 H&E 图像的深度学习自动区分神经内分泌癌和 3 级神经内分泌肿瘤。
神经内分泌肿瘤(NENs)来源于弥漫性神经内分泌细胞,分为分化良好、增殖较少的神经内分泌肿瘤(NETs)(分为低级(G1)、中级(G2)和高级(G3)),或分化不良、增殖较多的神经内分泌癌(NECs)。低级别神经内分泌癌通常需要手术治疗,而高级别神经内分泌癌通常需要化疗。不过,低级别 NEN 可能具有侵袭性。因此,精确细化 NEN 的诊断至关重要。当分化/非分化明显或 Ki-67 或有丝分裂指数较低时,就可以进行精确诊断。对于形态上未分化但 Ki-67 百分比高和(或)有丝分裂指数高的病例,我们面临着挑战。为了应对这一挑战,我们开发了一种名为 NEToC 的深度学习(DL)系统,旨在完全利用免疫组化图像中的形态学信息来区分 NET 和 NEC,而不依赖于 Ki-67 或有丝分裂的评估。NEToC 的开发使用了西班牙 Parc Tauli 医院 2015 年至 2018 年期间的 95 个 NEN 病例,包括 588 张图像。NEToC以图形用户界面(GUI)系统的形式实施,旨在部署到医院的病理科,以执行联合监督。我们使用人工神经网络(ANN)训练阶段未使用的 119 幅图像测试了 NEToC 的性能,并评估了其在不同分辨率下的鲁棒性:64 × 64、128 × 128、256 × 256 和 512 × 512 像素。对于代表性不足的 NET G3 实验,这些分辨率的准确率分别为 74%、98%、98% 和 100%;对于代表性较强的 NET G3 实验,准确率分别为 66%、89%、95% 和 94%。根据几个测得的性能指标,考虑到计算资源和精度要求,最佳分辨率似乎介于 128 × 128 和 256 × 256 像素之间。不过,我们发现 256 × 256 像素的分辨率在学习阶段对代表性不足的类别进行分类时更为稳健。这些结果表明,区分 NEC 和 3 级 NET 的信息需要在像素分辨率不超过 4 μm/pixel 的区域内解析。大多数错误分类都是假阴性,即 G1 型 NET 图像被错误地分类为 NEC 型。我们的结果表明,在医生面临挑战的 NEN 病例中,基于 DL 的诊断算法能提供更准确的诊断。NEToC 最初是用胃肠道 NEN 进行训练和分类的。由于不同器官的 NEN 形态没有变化,NEToC 的使用可以推广到不同器官的 NEN。NEToC 为联合监督提供了便利,病理学家可以根据 NEToC 的分类预测收集可互换的文件。NEToC 是一款易于使用、适应性强的软件,它集成了多个 ANN,可提高 NEN 诊断的标准化和准确性,为在联合监督系统中结合 DL 和组织学诊断提供了可能性。未来的目标是不仅对 NET 进行分类,还能完全根据组织分化信息对三级系统(NET G1、NET G2 和 NET G3)进行分类。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


