Identification and validation of miR-21 key genes in cervical cancer through an integrated bioinformatics approach
IF 3.1
4区 生物学
Q2 BIOLOGY
引用次数: 0
Abstract
Cervical cancer is one of the most prevalent female reproductive cancers. miR-21 is a multi-target oncomiR that has shown its potential in regulating several cancers including colon, pancreatic, breast, prostate, ovarian, and cervical cancer. However, the signaling network of miR-21 remains underexplored, and only a limited number of miR-21 gene targets in cervical cancer have been reported. In this context, the present study was undertaken to evaluate the role of miR-21 in cervical cancer by combining in silico analysis with in vitro validation in cervical cancer cells. The miR-21 target genes were predicted using four different prediction tools: miRWalk, DIANA, miRDB, and TargetScan. A total of 113 overlapping target genes, common in at least three of the prediction tools, were shortlisted and subjected to functional enrichment analysis. The analysis predicted that JAK-STAT, MAPK, neurotrophin, and Ras signaling pathways are significantly (p≤0.05) targeted by miR-21. The MCODE plugin identified the potential cluster in the protein-protein interaction network based on the highest degree of connectivity. After GEPIA2 validation of all 20 hub genes, NTF3, LIFR, and IL-6R were shortlisted for validation in cervical cancer cell lines. The results showed that NTF3, LIFR, and IL-6R were significantly upregulated in the miR-21 knockdown CaSki cell lines in 6.27, 1.92 and 1.71 folds (p≤0.01), respectively. Similarly, in HeLa cell lines expression of NTF3, LIFR, and IL-6R were overexpressed in 4.06, 5.65, 2.42 folds (p≤0.001), respectively. Findings of the study was confirming the role of miR-21 in regulating the expression of these genes. Additionally, the knockdown of miR-21 significantly inhibited the secretion of matrix metalloproteinases by CaSki cells. These results highlight that miR-21 could be a potential therapeutic target for cervical cancer, although further preclinical and clinical studies are required to validate its role and efficacy.
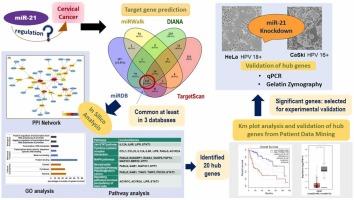
通过综合生物信息学方法鉴定和验证宫颈癌中的 miR-21 关键基因。
宫颈癌是最常见的女性生殖系统癌症之一。miR-21 是一种多靶点的癌基因,已显示出它在调节包括结肠癌、胰腺癌、乳腺癌、前列腺癌、卵巢癌和宫颈癌在内的多种癌症方面的潜力。然而,miR-21 的信号转导网络仍未得到充分探索,宫颈癌中的 miR-21 基因靶点也仅有少量报道。在此背景下,本研究通过结合硅学分析和宫颈癌细胞的体外验证,评估了 miR-21 在宫颈癌中的作用。研究人员使用四种不同的预测工具:miRWalk、DIANA、miRDB 和 TargetScan 预测了 miR-21 的靶基因。共筛选出 113 个重叠的靶基因,这些基因在至少三种预测工具中都是常见的,并对其进行了功能富集分析。分析预测,miR-21 显著靶向 JAK-STAT、MAPK、神经营养素和 Ras 信号通路(p≤0.05)。MCODE 插件根据连接度最高的蛋白质-蛋白质相互作用网络确定了潜在的群集。在对所有20个中心基因进行GEPIA2验证后,NTF3、LIFR和IL-6R入围宫颈癌细胞系的验证。结果显示,在 miR-21 敲除的 CaSki 细胞系中,NTF3、LIFR 和 IL-6R 分别显著上调了 6.27、1.92 和 1.71 倍(p≤0.01)。同样,在 HeLa 细胞系中,NTF3、LIFR 和 IL-6R 的表达分别过表达了 4.06、5.65 和 2.42 倍(p≤0.001)。研究结果证实了 miR-21 在调节这些基因表达中的作用。此外,敲除 miR-21 能显著抑制 CaSki 细胞分泌基质金属蛋白酶。这些结果突出表明,miR-21 可能是宫颈癌的潜在治疗靶点,尽管还需要进一步的临床前和临床研究来验证其作用和疗效。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


