Covalent DNA-Encoded Library Workflow Drives Discovery of SARS-CoV-2 Nonstructural Protein Inhibitors
IF 14.4
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
The COVID-19 pandemic, exacerbated by persistent viral mutations, underscored the urgent need for diverse inhibitors targeting multiple viral proteins. In this study, we utilized covalent DNA-encoded libraries to discover innovative triazine-based covalent inhibitors for the 3-chymotrypsin-like protease (3CLpro, Nsp5) and the papain-like protease (PLpro) domains of Nsp3, as well as novel non-nucleoside covalent inhibitors for the nonstructural protein 12 (Nsp12, RdRp). Optimization through molecular docking and medicinal chemistry led to the development of LU9, a nonpeptide 3CLpro inhibitor with an IC50 of 0.34 μM, and LU10, whose crystal structure showed a distinct binding mode within the 3CLpro active site. The X-ray cocrystal structure of SARS-CoV-2 PLpro in complex with XD5 uncovered a previously unexplored binding site adjacent to the catalytic pocket. Additionally, a non-nucleoside covalent Nsp12 inhibitor XJ5 achieved a potency of 0.12 μM following comprehensive structure–activity relationship analysis and optimization. Molecular dynamics revealed a potential binding mode. These compounds offer valuable chemical probes for target validation and represent promising candidates for the development of SARS-CoV-2 antiviral therapies.
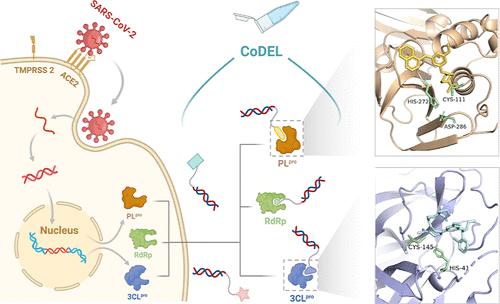
共价 DNA 编码库工作流程推动了 SARS-CoV-2 非结构蛋白抑制剂的发现
COVID-19大流行因病毒的持续突变而加剧,这凸显了对针对多种病毒蛋白的多样化抑制剂的迫切需求。在这项研究中,我们利用共价 DNA 编码文库发现了创新的三嗪类共价抑制剂,用于抑制 Nsp3 的 3-糜蛋白酶样蛋白酶(3CLpro,Nsp5)和木瓜蛋白酶样蛋白酶(PLpro)结构域,以及非结构蛋白 12(Nsp12,RdRp)的新型非核苷类共价抑制剂。通过分子对接和药物化学的优化,开发出了 LU9(一种 IC50 为 0.34 μM 的非肽 3CLpro 抑制剂)和 LU10(其晶体结构显示了 3CLpro 活性位点内独特的结合模式)。SARS-CoV-2 PLpro 与 XD5 复合物的 X 射线共晶体结构发现了一个以前未曾探索过的与催化口袋相邻的结合位点。此外,经过全面的结构-活性关系分析和优化,一种非核苷共价 Nsp12 抑制剂 XJ5 的效力达到了 0.12 μM。分子动力学揭示了一种潜在的结合模式。这些化合物为靶点验证提供了宝贵的化学探针,是开发 SARS-CoV-2 抗病毒疗法的有希望的候选化合物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
24.40
自引率
6.00%
发文量
2398
审稿时长
1.6 months
期刊介绍:
The flagship journal of the American Chemical Society, known as the Journal of the American Chemical Society (JACS), has been a prestigious publication since its establishment in 1879. It holds a preeminent position in the field of chemistry and related interdisciplinary sciences. JACS is committed to disseminating cutting-edge research papers, covering a wide range of topics, and encompasses approximately 19,000 pages of Articles, Communications, and Perspectives annually. With a weekly publication frequency, JACS plays a vital role in advancing the field of chemistry by providing essential research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


