Iris Fañanás-Pueyo, Ana-Mariya Anhel, Ángel Goñi-Moreno, Luis Oñate-Sánchez, Gerardo Carrera-Castaño
{"title":"Workflow to Select Functional Promoter DNA Baits and Screen Arrayed Gene Libraries in Yeast","authors":"Iris Fañanás-Pueyo, Ana-Mariya Anhel, Ángel Goñi-Moreno, Luis Oñate-Sánchez, Gerardo Carrera-Castaño","doi":"10.1002/cpz1.70059","DOIUrl":null,"url":null,"abstract":"<p>The yeast one-hybrid system (Y1H) is used extensively to identify DNA–protein interactions. The generation of large collections of open reading frames (ORFs) to be used as prey in screenings is not a bottleneck nowadays and can be carried out in-house or offered as a service by companies. However, the straightforward use of full gene promoters as baits to identify interacting proteins undermines the accuracy and sensitivity of the assay, especially in the case of multicellular eukaryotes. Therefore, it is paramount to implement procedures for efficient identification of suitable promoter fragments compatible with the Y1H assay. Here, we describe a workflow to identify biologically relevant conserved promoter fragments of <i>Arabidopsis thaliana</i> through simple and robust phylogenetic analyses. Additionally, we describe a manual method and its automated robotized version for rapid and efficient high-throughput Y1H screenings of arrayed ORF libraries with the identified DNA fragments. Moreover, this method can be scaled up or down and used for yeast two-hybrid screenings to search for possible interactors of proteins identified by the Y1H approach or any other protein of interest, altogether underscoring its suitability to build gene regulatory networks. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.</p><p><b>Basic Protocol 1</b>: Selection of DNA baits for Y1H screenings</p><p><b>Basic Protocol 2</b>: Y1H screenings with arrayed gene libraries</p><p><b>Alternate Protocol</b>: Automated screening with a liquid-handling robot</p>","PeriodicalId":93970,"journal":{"name":"Current protocols","volume":"4 11","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2024-11-21","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/cpz1.70059","citationCount":"0","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpz1.70059","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 0
Abstract
The yeast one-hybrid system (Y1H) is used extensively to identify DNA–protein interactions. The generation of large collections of open reading frames (ORFs) to be used as prey in screenings is not a bottleneck nowadays and can be carried out in-house or offered as a service by companies. However, the straightforward use of full gene promoters as baits to identify interacting proteins undermines the accuracy and sensitivity of the assay, especially in the case of multicellular eukaryotes. Therefore, it is paramount to implement procedures for efficient identification of suitable promoter fragments compatible with the Y1H assay. Here, we describe a workflow to identify biologically relevant conserved promoter fragments of Arabidopsis thaliana through simple and robust phylogenetic analyses. Additionally, we describe a manual method and its automated robotized version for rapid and efficient high-throughput Y1H screenings of arrayed ORF libraries with the identified DNA fragments. Moreover, this method can be scaled up or down and used for yeast two-hybrid screenings to search for possible interactors of proteins identified by the Y1H approach or any other protein of interest, altogether underscoring its suitability to build gene regulatory networks. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: Selection of DNA baits for Y1H screenings
Basic Protocol 2: Y1H screenings with arrayed gene libraries
Alternate Protocol: Automated screening with a liquid-handling robot
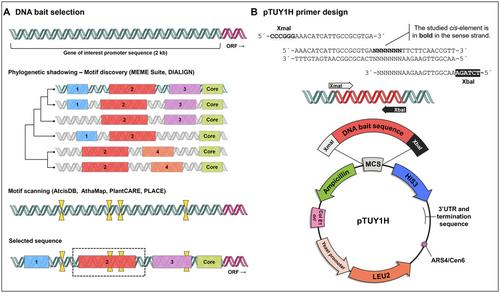
在酵母中选择功能性启动子 DNA 诱饵和筛选阵列基因库的工作流程。
酵母单杂交系统(Y1H)被广泛用于鉴定 DNA 蛋白相互作用。如今,生成大量开放阅读框(ORF)作为筛选的猎物已不再是瓶颈,可以在公司内部进行,也可以由公司提供服务。然而,直接使用全基因启动子作为诱饵来鉴定相互作用的蛋白质会降低检测的准确性和灵敏度,尤其是在多细胞真核生物中。因此,最重要的是实施有效识别与 Y1H 检测兼容的合适启动子片段的程序。在这里,我们介绍了一种通过简单而稳健的系统发育分析鉴定拟南芥生物学相关保守启动子片段的工作流程。此外,我们还介绍了一种手动方法及其自动化机器人版本,该方法可快速高效地利用鉴定出的 DNA 片段对阵列 ORF 文库进行高通量 Y1H 筛选。此外,这种方法还可以放大或缩小,并用于酵母双杂交筛选,以寻找通过 Y1H 方法鉴定的蛋白质或任何其他感兴趣的蛋白质的可能相互作用者,从而突出了它在构建基因调控网络方面的适用性。© 2024 作者。当前协议》由 Wiley Periodicals LLC 出版。基本方案 1:为 Y1H 筛选选择 DNA 诱饵 基本方案 2:使用阵列基因库进行 Y1H 筛选 替代方案:使用液体处理机器人进行自动筛选。
本文章由计算机程序翻译,如有差异,请以英文原文为准。


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


