Molecular heterogeneity in urothelial carcinoma and determinants of clinical benefit to PD-L1 blockade
IF 48.8
1区 医学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Checkpoint inhibitors targeting programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) have revolutionized cancer therapy across many indications including urothelial carcinoma (UC). Because many patients do not benefit, a better understanding of the molecular mechanisms underlying response and resistance is needed to improve outcomes. We profiled tumors from 2,803 UC patients from four late-stage randomized clinical trials evaluating the PD-L1 inhibitor atezolizumab by RNA sequencing (RNA-seq), a targeted DNA panel, immunohistochemistry, and digital pathology. Machine learning identifies four transcriptional subtypes, representing luminal desert, stromal, immune, and basal tumors. Overall survival benefit from atezolizumab over standard-of-care is observed in immune and basal tumors, through different response mechanisms. A self-supervised digital pathology approach can classify molecular subtypes from H&E slides with high accuracy, which could accelerate tumor molecular profiling. This study represents a large integration of UC molecular and clinical data in randomized trials, paving the way for clinical studies tailoring treatment to specific molecular subtypes in UC and other indications.
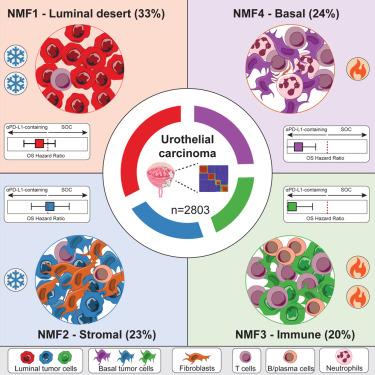
尿路上皮癌的分子异质性及 PD-L1 阻断剂临床获益的决定因素
以程序性细胞死亡蛋白1(PD-1)/程序性死亡配体1(PD-L1)为靶点的检查点抑制剂彻底改变了包括尿路上皮癌(UC)在内的多种适应症的癌症治疗。由于许多患者无法从中获益,因此需要更好地了解反应和耐药性的分子机制,以改善治疗效果。我们通过RNA测序(RNA-seq)、靶向DNA面板、免疫组化和数字病理学,对来自四项评估PD-L1抑制剂atezolizumab的晚期随机临床试验的2803名UC患者的肿瘤进行了分析。机器学习确定了四种转录亚型,分别代表管腔荒漠型、基质型、免疫型和基底型肿瘤。通过不同的反应机制,在免疫性肿瘤和基底层肿瘤中观察到阿特珠单抗的总体生存期优于标准疗法。自我监督数字病理学方法能从 H&E 切片中高精度地划分分子亚型,从而加快肿瘤分子图谱的绘制。这项研究代表了随机试验中 UC 分子和临床数据的大规模整合,为针对 UC 和其他适应症的特定分子亚型进行治疗的临床研究铺平了道路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cancer Cell
医学-肿瘤学
CiteScore
55.20
自引率
1.20%
发文量
179
审稿时长
4-8 weeks
期刊介绍:
Cancer Cell is a journal that focuses on promoting major advances in cancer research and oncology. The primary criteria for considering manuscripts are as follows:
Major advances: Manuscripts should provide significant advancements in answering important questions related to naturally occurring cancers.
Translational research: The journal welcomes translational research, which involves the application of basic scientific findings to human health and clinical practice.
Clinical investigations: Cancer Cell is interested in publishing clinical investigations that contribute to establishing new paradigms in the treatment, diagnosis, or prevention of cancers.
Insights into cancer biology: The journal values clinical investigations that provide important insights into cancer biology beyond what has been revealed by preclinical studies.
Mechanism-based proof-of-principle studies: Cancer Cell encourages the publication of mechanism-based proof-of-principle clinical studies, which demonstrate the feasibility of a specific therapeutic approach or diagnostic test.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


