Identification of molecular and cellular infection response biomarkers associated with anthrax infection through comparative analysis of gene expression data
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
Abstract
Bacillus anthracis, a gram-positive bacillus capable of forming spores, causes anthrax in mammals, including humans, and is recognized as a potential biological weapon agent. The diagnosis of anthrax is challenging due to variable symptoms resulting from exposure and infection severity. Despite the availability of a licensed vaccines, their limited long-term efficacy underscores the inadequacy of current human anthrax vaccines, highlighting the urgent need for next-generation alternatives. Our study aimed to identify molecular biomarkers and essential biological pathways for the early detection and accurate diagnosis of human anthrax infection. Using a comparative analysis of Bacillus anthracis gene expression data from the Gene Expression Omnibus (GEO) database, this cost-effective approach enables the identification of shared differentially expressed genes (DEGs) across separate microarray datasets without additional hybridization. Three microarray datasets (GSE34407, GSE14390, and GSE12131) of B. anthracis-infected human cell lines were analyzed via the GEO2R tool to identify shared DEGs. We identified 241 common DEGs (70 upregulated and 171 downregulated) from cell lines treated similarly to lethal toxins. Additionally, 10 common DEGs (5 upregulated and 5 downregulated) were identified across different treatments (lethal toxins and spores) and cell lines. Network meta-analysis identified JUN and GATAD2A as the top hub genes for overexpression, and NEDD4L and GULP1 for underexpression. Furthermore, prognostic analysis and SNP detection of the two identified upregulated hub genes were carried out in conjunction with machine learning classification models, with SVM yielding the best classification accuracy of 87.5 %. Our comparative analysis of Bacillus anthracis infection revealed striking similarities in gene expression 241 profiles across diverse datasets, despite variations in treatments and cell lines. These findings underscore how anthrax infection activates shared genes across different cell types, emphasizing this approach in the discovery of novel gene markers. These markers offer insights into pathogenesis and may lead to more effective therapeutic strategies. By identifying these genetic indicators, we can advance the development of precise immunotherapies, potentially enhancing vaccine efficacy and treatment outcomes.
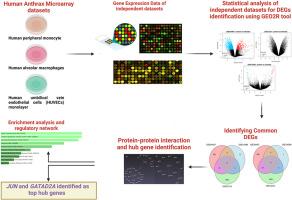
通过基因表达数据的比较分析,确定与炭疽感染相关的分子和细胞感染反应生物标志物
炭疽杆菌是一种能形成孢子的革兰氏阳性杆菌,可导致包括人类在内的哺乳动物患炭疽病,被认为是一种潜在的生物武器病原体。由于接触和感染严重程度不同导致的症状各异,因此炭疽的诊断具有挑战性。尽管目前已有获得许可的疫苗,但其有限的长期疗效凸显了目前人类炭疽疫苗的不足,突出表明了对下一代替代疫苗的迫切需求。我们的研究旨在确定早期检测和准确诊断人类炭疽感染的分子生物标志物和重要生物途径。通过对基因表达总库(GEO)数据库中的炭疽杆菌基因表达数据进行比较分析,这种经济有效的方法无需额外的杂交,就能在不同的微阵列数据集之间识别共有的差异表达基因(DEGs)。我们通过 GEO2R 工具分析了炭疽杆菌感染的人类细胞系的三个微阵列数据集(GSE34407、GSE14390 和 GSE12131),以确定共有的 DEGs。我们从受到致命毒素类似处理的细胞系中发现了 241 个共有 DEGs(70 个上调,171 个下调)。此外,我们还在不同的处理(致死毒素和孢子)和细胞系中发现了 10 个共同的 DEGs(5 个上调,5 个下调)。网络荟萃分析发现 JUN 和 GATAD2A 是表达过高的首要枢纽基因,而 NEDD4L 和 GULP1 则是表达过低的首要枢纽基因。此外,我们还结合机器学习分类模型对这两个上调的枢纽基因进行了预后分析和SNP检测,其中SVM的分类准确率最高,达到87.5%。我们对炭疽杆菌感染的比较分析表明,尽管治疗方法和细胞系不同,但不同数据集的基因表达241图谱具有惊人的相似性。这些发现强调了炭疽感染如何激活不同细胞类型中的共享基因,并强调了发现新型基因标记物的方法。这些标记可帮助我们深入了解致病机理,并可能带来更有效的治疗策略。通过确定这些基因指标,我们可以推进精确免疫疗法的开发,从而有可能提高疫苗疗效和治疗效果。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


