Pseudo-label guided selective mutual learning for semi-supervised 3D medical image segmentation
IF 4.9
2区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
Abstract
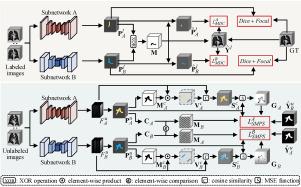
Semi-supervised learning (SSL) have shown promising results in 3D medical image segmentation by utilizing both labeled and readily available unlabeled images. Most current SSL methods predict unlabeled data under different perturbations by employing subnetworks with same architecture. Despite their progress, the homogenization of subnetworks limits the diverse predictions on both labeled and unlabeled data, thereby making it difficult for subnetworks to correct each other and giving rise to confirmation bias issue. In this paper, we introduce an SSL framework termed pseudo-label guided selective mutual learning (PLSML), which incorporates two distinct subnetworks and selectively utilizes their derived pseudo-labels for mutual supervision to mitigate the above issue. Specifically, the discrepancies of pseudo-labels from two distinct subnetworks are used to select the regions within labeled images that are prone to missegmentation. We then introduce a mutual discrepancy correction (MDC) regularization to revisit these regions. Moreover, a selective mutual pseudo supervision (SMPS) regularization is introduced to estimate the reliability of pseudo-labels of unlabeled images, and selectively leverage the more reliable pseudo-labels in the two subnetworks to supervise the other one. The integration of MDC and SMPS regularizations facilitates inter-subnetwork mutual correction, consequently mitigating confirmation bias. Extensive experiments on two 3D medical image datasets demonstrate the superiority of our PLSML as compared to state-of-the-art SSL methods. The source code is available online at https://github.com/1pca0/PLSML.
用于半监督三维医学图像分割的伪标签引导选择性相互学习
半监督学习(SSL)在三维医学图像分割方面取得了可喜的成果,它既能利用已标记的图像,也能利用随时可用的未标记图像。目前大多数半监督学习方法都是通过采用相同架构的子网络来预测不同扰动下的非标记数据。尽管这些方法取得了进步,但子网络的同质化限制了对有标签和无标签数据的不同预测,从而使子网络难以相互修正,并引发确认偏差问题。在本文中,我们介绍了一种名为伪标签引导的选择性相互学习(PLSML)的 SSL 框架,它包含两个不同的子网络,并选择性地利用其衍生的伪标签进行相互监督,以缓解上述问题。具体来说,利用来自两个不同子网络的伪标签的差异来选择标签图像中容易发生误判的区域。然后,我们引入相互差异校正(MDC)正则化来重新审视这些区域。此外,我们还引入了选择性相互伪监督(SMPS)正则化来估计未标注图像的伪标签的可靠性,并选择性地利用两个子网络中更可靠的伪标签来监督另一个子网络。MDC 和 SMPS 正则化的整合促进了子网络间的相互校正,从而减轻了确认偏差。在两个三维医学图像数据集上进行的大量实验证明,与最先进的 SSL 方法相比,我们的 PLSML 更具优势。源代码可从 https://github.com/1pca0/PLSML 在线获取。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Biomedical Signal Processing and Control
工程技术-工程:生物医学
CiteScore
9.80
自引率
13.70%
发文量
822
审稿时长
4 months
期刊介绍:
Biomedical Signal Processing and Control aims to provide a cross-disciplinary international forum for the interchange of information on research in the measurement and analysis of signals and images in clinical medicine and the biological sciences. Emphasis is placed on contributions dealing with the practical, applications-led research on the use of methods and devices in clinical diagnosis, patient monitoring and management.
Biomedical Signal Processing and Control reflects the main areas in which these methods are being used and developed at the interface of both engineering and clinical science. The scope of the journal is defined to include relevant review papers, technical notes, short communications and letters. Tutorial papers and special issues will also be published.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


