Quantification of bacterial shape using moment invariants enables distinguishing populations during cellular plasmolysis
IF 1.6
Q2 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
The analysis of geometrical cell shape is fundamental to understand motility, development, and responses to external stimuli. The moment invariants framework quantifies cellular shape and size, although its applicability has not been explored for rod-shaped bacteria. In this work, we use moment invariants to evaluate the extent of cell shape change (projected area and volume) during plasmolysis, as Escherichia coli cells are subjected to hyperosmotic shock. The characteristic cell size descriptors width, length and area show systematic decrease as external salt (NaCl) conditions increase—except for high salt, where a small population of cells shows evidence of membrane rupture. We use these two-dimensional results to estimate cell volume during plasmolysis, finding a minimum volume that is not reduced further with increase in salt concentration. Next, we computed elongation and dispersion, metrics that quantify how cell shape is stretched out or differs from an ellipse, respectively. For dispersion, we observe the development of a long tail for the distribution at high salt. Moreover, the use of elongation-dispersion plots enables distinction of plasmolyzed and normal cells despite the presence of broad distributions. Altogether, a protocol is provided to evaluate bacterial shape, highlighting a set of metrics that help distinguish among bacterial populations.
- •Moment invariants enable quantitative description of bacterial morphology in two dimensions, and estimation of volume
- •We apply the moment invariants framework to describe changes in bacterial shape during plasmolysis
- •The proposed methodology shows suitability to distinguish among cellular populations.
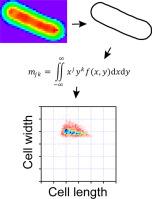
利用矩不变式量化细菌形状,可在细胞解痉过程中区分菌群
分析细胞的几何形状是了解细胞运动、发育和对外部刺激做出反应的基础。矩不变式框架可量化细胞的形状和大小,但其对杆状细菌的适用性尚未得到探讨。在这项研究中,我们利用力矩不变式来评估大肠杆菌细胞在受到高渗冲击时,在质解过程中细胞形状变化的程度(投影面积和体积)。随着外部盐分(NaCl)条件的增加,特征细胞尺寸描述符的宽度、长度和面积显示出系统性的减少--除了在高盐分条件下,一小部分细胞显示出膜破裂的迹象。我们利用这些二维结果来估计解痉过程中的细胞体积,发现了一个最小体积,该体积不会随着盐浓度的增加而进一步缩小。接下来,我们计算了伸长率和离散度,这两个指标分别量化了细胞形状的伸长程度或与椭圆的差异。在离散度方面,我们观察到在高盐条件下分布出现了长尾。此外,尽管存在宽广的分布,但使用伸长-离散图可以区分解痉细胞和正常细胞。我们应用矩不变式框架来描述解痉过程中细菌形状的变化。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

MethodsX
Health Professions-Medical Laboratory Technology
CiteScore
3.60
自引率
5.30%
发文量
314
审稿时长
7 weeks
期刊介绍:
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


