Validation of reference genes for cardiac RT-qPCR studies spanning the fetal to adult period
IF 1.9
Q2 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Many genes used as internal controls for mRNA expression studies are unstable (change) over development. This study determined an approach to validate reference genes for mRNA studies spanning the fetal period to adulthood in sheep hearts.
- •We determined the mRNA expression of 12 candidate reference genes (ACTB, GAPDH, H3-3A, HYAL2, PPIA, RNA18S1, RPL32, RPL37A, RPL41, RPLP0, RPS15, and YWHAZ) via RT-qPCR. Per RefFinder, which incorporates computational algorithms by BestKeeper, comparative delta Ct, GeNorm, and NormFinder, RPL32, RPL37A, HYAL2, ACTB and GAPDH were the most stable reference genes, although none were unchanged across all ages.
- •Systematical calculation of the geometric means of 3 reference genes revealed the combination of HYAL2, RPL32, and RPL37A was unchanged across the 5 fetal, neonatal, and adult ages.
- •We determined the most stable combination of reference genes for cardiac gene expression studies in sheep from fetus to newborn to adult; these steps are applicable to determine internal controls for mRNA studies in other organs, other species, and periods in which reference gene instability is high.
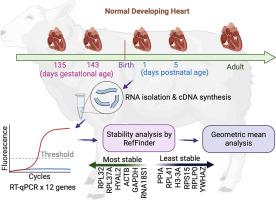
验证从胎儿期到成年期心脏 RT-qPCR 研究的参考基因
许多用作 mRNA 表达研究内部对照的基因在发育过程中并不稳定(会发生变化)。我们通过 RT-qPCR 确定了 12 个候选参考基因(ACTB、GAPDH、H3-3A、HYAL2、PPIA、RNA18S1、RPL32、RPL37A、RPL41、RPLP0、RPS15 和 YWHAZ)的 mRNA 表达。根据结合了 BestKeeper、比较 delta Ct、GeNorm 和 NormFinder 等计算算法的 RefFinder,RPL32、RPL37A、HYAL2、ACTB 和 GAPDH 是最稳定的参考基因,尽管它们在所有年龄段都没有变化。-我们确定了绵羊从胎儿到新生儿再到成年的心脏基因表达研究中最稳定的参考基因组合;这些步骤适用于确定其他器官、其他物种以及参考基因不稳定性较高的时期的 mRNA 研究的内部对照。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

MethodsX
Health Professions-Medical Laboratory Technology
CiteScore
3.60
自引率
5.30%
发文量
314
审稿时长
7 weeks
期刊介绍:
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


