Review and revamp of compositional data transformation: A new framework combining proportion conversion and contrast transformation
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
Computational and structural biotechnology journal
Pub Date : 2024-11-08
DOI:10.1016/j.csbj.2024.11.003
引用次数: 0
Abstract
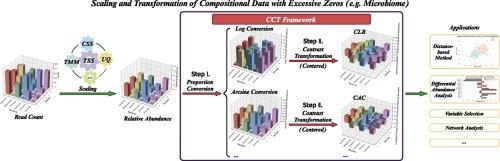
Due to the development of next-generation sequencing technology and an increased appreciation of their role in modulating host immunity and their potential as therapeutic agents, the human microbiome has emerged as a key area of interest in various biological investigations of human health and disease. However, microbiome data present a number of statistical challenges not addressed by existing methods, such as the varying sequencing depth, the compositionality, and zero inflation. Solutions like scaling and transformation methods help to mitigate heterogeneity and release constraints, but often introduce biases and yield inconsistent results on the same data. To address these issues, we conduct a systematic review of compositional data transformation, with a particular focus on the connection and distinction of existing techniques. Additionally, we create a new framework that enables the development of new transformations by combining proportion conversion with contrast transformations. This framework includes well-known methods such as Additive Log Ratio (ALR) and Centered Log Ratio (CLR) as special cases. Using this framework, we develop two novel transformations—Centered Arcsine Contrast (CAC) and Additive Arcsine Contrast (AAC)—which show enhanced performance in scenarios with high zero-inflation. Moreover, our findings suggest that ALR and CLR transformations are more effective when zero values are less prevalent. This comprehensive review and the innovative framework provide microbiome researchers with a significant direction to enhance data transformation procedures and improve analytical outcomes.
审查和修改合成数据转换:比例转换与对比度转换相结合的新框架
由于下一代测序技术的发展,以及人们对微生物在调节宿主免疫力中的作用及其作为治疗药物的潜力的认识不断提高,人类微生物组已成为人类健康和疾病的各种生物学研究中的一个关键领域。然而,微生物组数据存在许多现有方法无法解决的统计难题,如不同的测序深度、组成和零膨胀。缩放和转换方法等解决方案有助于缓解异质性和释放限制,但往往会带来偏差,并在相同数据上产生不一致的结果。为了解决这些问题,我们对成分数据转换进行了系统回顾,尤其关注现有技术之间的联系和区别。此外,我们还创建了一个新的框架,通过将比例转换与对比度转换相结合来开发新的转换方法。这个框架包括众所周知的方法,如作为特例的加法对数比(ALR)和居中对数比(CLR)。利用这一框架,我们开发了两种新型转换--居中弧线对比度 (CAC) 和加性弧线对比度 (AAC),这两种转换在零膨胀率较高的情况下表现出更强的性能。此外,我们的研究结果表明,当零值不那么普遍时,ALR 和 CLR 变换更为有效。这篇综述和创新框架为微生物组研究人员提供了一个重要的方向,以加强数据转换程序并改善分析结果。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational and structural biotechnology journal
Biochemistry, Genetics and Molecular Biology-Biophysics
CiteScore
9.30
自引率
3.30%
发文量
540
审稿时长
6 weeks
期刊介绍:
Computational and Structural Biotechnology Journal (CSBJ) is an online gold open access journal publishing research articles and reviews after full peer review. All articles are published, without barriers to access, immediately upon acceptance. The journal places a strong emphasis on functional and mechanistic understanding of how molecular components in a biological process work together through the application of computational methods. Structural data may provide such insights, but they are not a pre-requisite for publication in the journal. Specific areas of interest include, but are not limited to:
Structure and function of proteins, nucleic acids and other macromolecules
Structure and function of multi-component complexes
Protein folding, processing and degradation
Enzymology
Computational and structural studies of plant systems
Microbial Informatics
Genomics
Proteomics
Metabolomics
Algorithms and Hypothesis in Bioinformatics
Mathematical and Theoretical Biology
Computational Chemistry and Drug Discovery
Microscopy and Molecular Imaging
Nanotechnology
Systems and Synthetic Biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


