A review on federated learning in computational pathology
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
Computational and structural biotechnology journal
Pub Date : 2024-10-29
DOI:10.1016/j.csbj.2024.10.037
引用次数: 0
Abstract
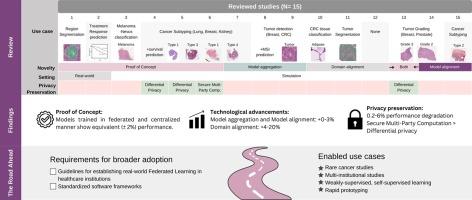
Training generalizable computational pathology (CPATH) algorithms is heavily dependent on large-scale, multi-institutional data. Simultaneously, healthcare data underlies strict data privacy rules, hindering the creation of large datasets. Federated Learning (FL) is a paradigm addressing this dilemma, by allowing separate institutions to collaborate in a training process while keeping each institution's data private and exchanging model parameters instead. In this study, we identify and review key developments of FL for CPATH applications. We consider 15 studies, thereby evaluating the current status of exploring and adapting this emerging technology for CPATH applications. Proof-of-concept studies have been conducted across a wide range of CPATH use cases, showcasing the performance equivalency of models trained in a federated compared to a centralized manner. Six studies focus on model aggregation or model alignment methods reporting minor () performance improvement compared to conventional FL techniques, while four studies explore domain alignment methods, resulting in more significant performance improvements (). To further reduce the privacy risk posed by sharing model parameters, four studies investigated the use of privacy preservation methods, where all methods demonstrated equivalent or slightly degraded performance ( lower). To facilitate broader, real-world environment adoption, it is imperative to establish guidelines for the setup and deployment of FL infrastructure, alongside the promotion of standardized software frameworks. These steps are crucial to 1) further democratize CPATH research by allowing smaller institutions to pool data and computational resources 2) investigating rare diseases, 3) conducting multi-institutional studies, and 4) allowing rapid prototyping on private data.
计算病理学中的联合学习综述
可通用计算病理学(CPATH)算法的训练在很大程度上依赖于大规模、多机构数据。同时,医疗保健数据受到严格的数据隐私规定的限制,阻碍了大型数据集的创建。联合学习(FL)是解决这一难题的一种模式,它允许不同的机构在训练过程中进行合作,同时保持每个机构数据的私密性并交换模型参数。在本研究中,我们确定并回顾了 FL 在 CPATH 应用中的主要发展。我们考虑了 15 项研究,从而评估了在 CPATH 应用中探索和调整这一新兴技术的现状。我们对 CPATH 的各种用例进行了概念验证研究,展示了以联合方式训练的模型与集中方式训练的模型的性能等同性。六项研究重点关注模型聚合或模型对齐方法,结果表明与传统的 FL 技术相比,性能提高幅度较小(0∼3%),而四项研究探索了领域对齐方法,结果表明性能提高幅度更大(4∼20%)。为了进一步降低共享模型参数带来的隐私风险,四项研究调查了隐私保护方法的使用情况,所有方法都表现出同等或轻微的性能下降(低 0.2∼6%)。为了促进更广泛的真实环境应用,在推广标准化软件框架的同时,必须制定 FL 基础设施的设置和部署指南。这些步骤对以下方面至关重要:1)通过允许较小机构汇集数据和计算资源,进一步实现 CPATH 研究的民主化;2)调查罕见疾病;3)开展多机构研究;以及 4)允许在私人数据上快速建立原型。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational and structural biotechnology journal
Biochemistry, Genetics and Molecular Biology-Biophysics
CiteScore
9.30
自引率
3.30%
发文量
540
审稿时长
6 weeks
期刊介绍:
Computational and Structural Biotechnology Journal (CSBJ) is an online gold open access journal publishing research articles and reviews after full peer review. All articles are published, without barriers to access, immediately upon acceptance. The journal places a strong emphasis on functional and mechanistic understanding of how molecular components in a biological process work together through the application of computational methods. Structural data may provide such insights, but they are not a pre-requisite for publication in the journal. Specific areas of interest include, but are not limited to:
Structure and function of proteins, nucleic acids and other macromolecules
Structure and function of multi-component complexes
Protein folding, processing and degradation
Enzymology
Computational and structural studies of plant systems
Microbial Informatics
Genomics
Proteomics
Metabolomics
Algorithms and Hypothesis in Bioinformatics
Mathematical and Theoretical Biology
Computational Chemistry and Drug Discovery
Microscopy and Molecular Imaging
Nanotechnology
Systems and Synthetic Biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


