CL-MRI: Self-Supervised contrastive learning to improve the accuracy of undersampled MRI reconstruction
IF 4.9
2区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
Abstract
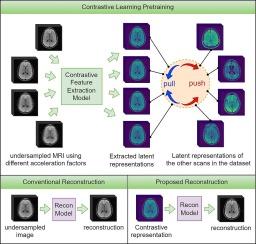
Deep learning (DL) methods have emerged as the state-of-the-art for Magnetic Resonance Imaging (MRI) reconstruction. DL methods typically involve training deep neural networks to take undersampled MRI images as input and transform them into high-quality MRI images through data-driven processes. However, deep learning models often fail with higher levels of undersampling due to the insufficient information in the input, which is crucial for producing high-quality MRI images. Thus, optimizing the information content at the input of a DL reconstruction model could significantly improve reconstruction accuracy. In this paper, we introduce a self-supervised pretraining procedure using contrastive learning to improve the accuracy of undersampled DL MRI reconstruction. We use contrastive learning to transform the MRI image representations into a latent space that maximizes mutual information among different undersampled representations and optimizes the information content at the input of the downstream DL reconstruction models. Our experiments demonstrate improved reconstruction accuracy across a range of acceleration factors and datasets, both quantitatively and qualitatively. Furthermore, our extended experiments validate the proposed framework’s robustness under adversarial conditions, such as measurement noise, different k-space sampling patterns, and pathological abnormalities, and also prove the transfer learning capabilities on MRI datasets with completely different anatomy. Additionally, we conducted experiments to visualize and analyze the properties of the proposed MRI contrastive learning latent space. Code available here.
CL-MRI:自我监督对比学习提高欠采样磁共振成像重建的准确性
深度学习(DL)方法已成为磁共振成像(MRI)重建的最先进方法。深度学习方法通常包括训练深度神经网络,将采样不足的磁共振成像图像作为输入,并通过数据驱动过程将其转换为高质量的磁共振成像图像。然而,深度学习模型在较高的欠采样水平下往往会失败,原因是输入信息不足,而这对生成高质量的 MRI 图像至关重要。因此,优化 DL 重建模型输入端的信息含量可以显著提高重建精度。在本文中,我们介绍了一种使用对比学习的自监督预训练程序,以提高欠采样 DL MRI 重建的准确性。我们利用对比学习将 MRI 图像表征转换为一个潜空间,该潜空间能最大化不同欠采样表征之间的互信息,并优化下游 DL 重建模型输入端的信息含量。我们的实验证明,在各种加速因子和数据集上,重建精度都有了定量和定性的提高。此外,我们的扩展实验还验证了所提出的框架在诸如测量噪声、不同 k 空间采样模式和病理异常等对抗条件下的鲁棒性,并证明了在解剖结构完全不同的 MRI 数据集上的迁移学习能力。此外,我们还进行了实验,对所提出的磁共振成像对比学习潜空间的特性进行了可视化分析。代码在此提供。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Biomedical Signal Processing and Control
工程技术-工程:生物医学
CiteScore
9.80
自引率
13.70%
发文量
822
审稿时长
4 months
期刊介绍:
Biomedical Signal Processing and Control aims to provide a cross-disciplinary international forum for the interchange of information on research in the measurement and analysis of signals and images in clinical medicine and the biological sciences. Emphasis is placed on contributions dealing with the practical, applications-led research on the use of methods and devices in clinical diagnosis, patient monitoring and management.
Biomedical Signal Processing and Control reflects the main areas in which these methods are being used and developed at the interface of both engineering and clinical science. The scope of the journal is defined to include relevant review papers, technical notes, short communications and letters. Tutorial papers and special issues will also be published.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


