Investigation of cold adaptation mechanisms by transcriptome analysis in the liver of yellowtail kingfish (Seriola aureovittata)
IF 2.2
2区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
Comparative Biochemistry and Physiology D-Genomics & Proteomics
Pub Date : 2024-11-10
DOI:10.1016/j.cbd.2024.101358
引用次数: 0
Abstract
Cold stress is an extreme environmental stressor that constrains the economic development of aquaculture. Yellowtail kingfish (Seriola aureovittata) is a commercially important fish species, but its molecular mechanisms in response to cold stress remain unknown. In this study, we investigated the transcriptional response of yellowtail kingfish liver to cold stress (10 °C) using RNA-sequencing analysis. We obtained 83.21 Gb of clean data from fish in the control group (0 h) and at 6, 12, and 24 h post-stimulation. A total of 2900 differentially expressed genes were identified from the comparison of the bioinformatic data from cold-stressed and control groups. Enrichment analysis suggested that protein processing, energy and lipid metabolism, signal transduction, and stress-induced cell cycle changes were highly involved during cold adaptation. Transport and utilization of fatty acids and cell cycle arrest were enhanced, whereas the rate of glycogen metabolism and protein biosynthesis were inhibited to maintain energy balance and normal fluidity of the cell membrane, thereby enhancing the tolerance of yellowtail kingfish to cold stress. Our study uncovered molecular pathways and key regulatory genes that are crucial for cold adaptation in yellowtail kingfish. These results provide new insights that could inform selective breeding programs aimed at enhancing cold resistance in aquaculture.
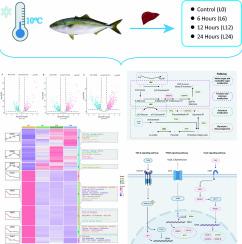
通过对黄尾鰤鱼(Seriola aureovittata)肝脏的转录组分析研究其冷适应机制。
冷胁迫是一种极端环境胁迫,制约着水产养殖业的经济发展。大黄鱼(Seriola aureovittata)是一种具有重要商业价值的鱼类,但其对冷胁迫的分子机制仍然未知。在这项研究中,我们利用 RNA 序列分析研究了大黄鱼肝脏对冷胁迫(10 °C)的转录响应。我们从对照组(0 h)和刺激后 6、12 和 24 h 的鱼体中获得了 83.21 Gb 的纯数据。通过比较冷应激组和对照组的生物信息学数据,共鉴定出 2900 个差异表达基因。富集分析表明,蛋白质加工、能量和脂质代谢、信号转导和应激诱导的细胞周期变化高度参与了冷适应过程。为了维持能量平衡和细胞膜的正常流动性,脂肪酸的运输和利用以及细胞周期的停滞都得到了加强,而糖原代谢和蛋白质的生物合成则受到了抑制,从而增强了黄尾鰤鱼对冷应激的耐受性。我们的研究发现了大黄鱼冷适应的分子途径和关键调控基因。这些结果为旨在提高水产养殖业抗寒能力的选择性育种计划提供了新的启示。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
5.10
自引率
3.30%
发文量
69
审稿时长
33 days
期刊介绍:
Comparative Biochemistry & Physiology (CBP) publishes papers in comparative, environmental and evolutionary physiology.
Part D: Genomics and Proteomics (CBPD), focuses on “omics” approaches to physiology, including comparative and functional genomics, metagenomics, transcriptomics, proteomics, metabolomics, and lipidomics. Most studies employ “omics” and/or system biology to test specific hypotheses about molecular and biochemical mechanisms underlying physiological responses to the environment. We encourage papers that address fundamental questions in comparative physiology and biochemistry rather than studies with a focus that is purely technical, methodological or descriptive in nature.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


