Positional encoding-guided transformer-based multiple instance learning for histopathology whole slide images classification
IF 4.9
2区 医学
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background and objectives:
Whole slide image (WSI) classification is of great clinical significance in computer-aided pathological diagnosis. Due to the high cost of manual annotation, weakly supervised WSI classification methods have gained more attention. As the most representative, multiple instance learning (MIL) generally aggregates the predictions or features of the patches within a WSI to achieve the slide-level classification under the weak supervision of WSI labels. However, most existing MIL methods ignore spatial position relationships of the patches, which is likely to strengthen the discriminative ability of WSI-level features.
Methods:
In this paper, we propose a novel positional encoding-guided transformer-based multiple instance learning (PEGTB-MIL) method for histopathology WSI classification. It aims to encode the spatial positional property of the patch into its corresponding semantic features and explore the potential correlation among the patches for improving the WSI classification performance. Concretely, the deep features of the patches in WSI are first extracted and simultaneously a position encoder is used to encode the spatial 2D positional information of the patches into the spatial-aware features. After incorporating the semantic features and spatial embeddings, multi-head self-attention (MHSA) is applied to explore the contextual and spatial dependencies of the fused features. Particularly, we introduce an auxiliary reconstruction task to enhance the spatial–semantic consistency and generalization ability of features.
Results:
The proposed method is evaluated on two public benchmark TCGA datasets (TCGA-LUNG and TCGA-BRCA) and two in-house clinical datasets (USTC-EGFR and USTC-GIST). Experimental results validate it is effective in the tasks of cancer subtyping and gene mutation status prediction. In the test stage, the proposed PEGTB-MIL outperforms the other state-of-the-art methods and respectively achieves 97.13±0.34%, 86.74±2.64%, 83.25±1.65%, and 72.52±1.63% of the area under the receiver operating characteristic (ROC) curve (AUC).
Conclusion:
PEGTB-MIL utilizes positional encoding to effectively guide and reinforce MIL, leading to enhanced performance on downstream WSI classification tasks. Specifically, the introduced auxiliary reconstruction module adeptly preserves the spatial–semantic consistency of patch features. More significantly, this study investigates the relationship between position information and disease diagnosis and presents a promising avenue for further research.
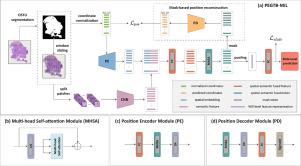
基于位置编码引导变换器的多实例学习,用于组织病理学整张切片图像分类。
背景和目的:全切片图像(WSI)分类在计算机辅助病理诊断中具有重要的临床意义。由于人工标注成本高昂,弱监督 WSI 分类方法受到越来越多的关注。最具代表性的是多实例学习(Multiple instance learning,MIL),一般是在 WSI 标签的弱监督下,将 WSI 中的斑块预测或特征汇总,实现玻片级分类。然而,现有的 MIL 方法大多忽略了斑块的空间位置关系,而这很可能会加强 WSI 级特征的判别能力:本文提出了一种用于组织病理学 WSI 分类的新型位置编码引导的基于变换器的多实例学习(PEGTB-MIL)方法。该方法旨在将斑块的空间位置属性编码为其相应的语义特征,并探索斑块之间潜在的相关性,以提高 WSI 分类性能。具体来说,首先提取 WSI 中补丁的深层特征,同时使用位置编码器将补丁的空间二维位置信息编码为空间感知特征。在整合语义特征和空间嵌入后,多头自注意(MHSA)将用于探索融合特征的上下文和空间依赖关系。特别是,我们引入了一项辅助重建任务,以增强特征的空间语义一致性和泛化能力:结果:我们在两个公共基准 TCGA 数据集(TCGA-LUNG 和 TCGA-BRCA)和两个内部临床数据集(USTC-EGFR 和 USTC-GIST)上对所提出的方法进行了评估。实验结果验证了它在癌症亚型划分和基因突变状态预测任务中的有效性。在测试阶段,所提出的 PEGTB-MIL 优于其他最先进的方法,分别达到了 97.13±0.34%、86.74±2.64%、83.25±1.65% 和 72.52±1.63% 的接收者操作特征曲线(ROC)下面积(AUC):结论:PEGTB-MIL 利用位置编码有效地指导和加强了 MIL,从而提高了下游 WSI 分类任务的性能。具体来说,引入的辅助重构模块能够很好地保留斑块特征的空间语义一致性。更重要的是,这项研究探讨了位置信息与疾病诊断之间的关系,为进一步的研究提供了一个前景广阔的途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computer methods and programs in biomedicine
工程技术-工程:生物医学
CiteScore
12.30
自引率
6.60%
发文量
601
审稿时长
135 days
期刊介绍:
To encourage the development of formal computing methods, and their application in biomedical research and medical practice, by illustration of fundamental principles in biomedical informatics research; to stimulate basic research into application software design; to report the state of research of biomedical information processing projects; to report new computer methodologies applied in biomedical areas; the eventual distribution of demonstrable software to avoid duplication of effort; to provide a forum for discussion and improvement of existing software; to optimize contact between national organizations and regional user groups by promoting an international exchange of information on formal methods, standards and software in biomedicine.
Computer Methods and Programs in Biomedicine covers computing methodology and software systems derived from computing science for implementation in all aspects of biomedical research and medical practice. It is designed to serve: biochemists; biologists; geneticists; immunologists; neuroscientists; pharmacologists; toxicologists; clinicians; epidemiologists; psychiatrists; psychologists; cardiologists; chemists; (radio)physicists; computer scientists; programmers and systems analysts; biomedical, clinical, electrical and other engineers; teachers of medical informatics and users of educational software.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


