Identification of significant hub genes and pathways associated with metastatic breast cancer and tolerogenic dendritic cell via bioinformatics analysis
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
Abstract
Metastatic breast cancer (MBC) is an advanced-stage breast cancer associated with more than 90 % of cancer-related deaths. Immunosuppressive properties of tolerogenic dendritic cells (tolDCs) in tumour immune microenvironment (TIME) may be a risk factor for the rapid progression to MBC. However, the exact connections between the two are unknown. The aim of the current study is to uncover gene signatures and key pathways associated with MBC and tolDCs via an integrated bioinformatics approach. Gene expression profiles of MBC and tolDCs were retrieved from Gene Expression Omnibus (GEO) to identify common differentially expressed genes (DEGs). From DGE analysis, 529 upregulated common DEGs and 367 downregulated common DEGs had been identified. In enrichment analysis, common DEGs enriched in GO terms of defense response to virus and KEGG pathway of transcriptional misregulation in cancer were reported to be significantly associated with MBC and tolDCs. From the constructed PPI networks, 23 hub genes were identified, although only 5 genes were significant; 3 upregulated (ISG15, OAS2 and RSAD2) and 2 downregulated (eEF2 and PPARG) as they were found to be significantly correlated and had the same expression trend as predicted in validation analysis of overall survival (OS) analysis, expression levels, immune infiltration analysis and immunohistochemistry (IHC) analysis. These 5 hub genes can now be exploited in developing novel therapeutic interventions and as diagnostic biomarkers for enhancing the clinical outcomes of MBC patients.
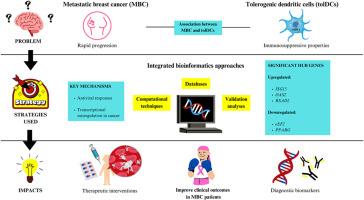
通过生物信息学分析,确定与转移性乳腺癌和耐受性树突状细胞相关的重要枢纽基因和通路。
转移性乳腺癌(MBC)是一种晚期乳腺癌,与超过 90% 的癌症相关死亡有关。肿瘤免疫微环境(TIME)中的耐受性树突状细胞(tolDCs)的免疫抑制特性可能是导致 MBC 快速发展的一个风险因素。然而,两者之间的确切联系尚不清楚。本研究旨在通过综合生物信息学方法发现与 MBC 和 tolDCs 相关的基因特征和关键通路。研究人员从基因表达总库(GEO)中检索了 MBC 和 tolDCs 的基因表达谱,以确定常见的差异表达基因(DEGs)。通过 DGE 分析,确定了 529 个上调的常见 DEGs 和 367 个下调的常见 DEGs。在富集分析中,富集在对病毒的防御反应的GO术语和癌症转录失调的KEGG通路中的常见DEGs被报道与MBC和tolDCs显著相关。从构建的 PPI 网络中,发现了 23 个枢纽基因,但只有 5 个基因是显著的;3 个上调(ISG15、OAS2 和 RSAD2)和 2 个下调(eEF2 和 PPARG),因为在总生存期(OS)分析、表达水平、免疫浸润分析和免疫组化(IHC)分析等验证分析中,发现这些基因与预测的表达趋势显著相关并具有相同的表达趋势。现在可以利用这5个枢纽基因开发新的治疗干预措施,并将其作为诊断生物标志物,以提高MBC患者的临床疗效。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


