Kinetics of i-motif folding within the duplex context
IF 2.2
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Non-canonical nucleic acid structures possess an ability to interact selectively with proteins, thereby exerting influence over various intracellular processes. Numerous studies indicate that genomic G-quadruplexes and i-motifs are involved in the regulation of transcription. These structures are formed temporarily during the unwinding of the DNA double helix; and their direct determination is a rather difficult task. In addition, i-motif folding is pH-dependent, with most i-motifs having low stability at neutral pH. However, some genomic i-motifs with long cytosine repeats were shown to be stable at pH 7.3, suggesting their functionality within the nucleus. Here we studied pH-dependent behavior of a model i-motif with flanking sequences that forms a duplex motif. Kinetic studies on bimodular structures with cytosine residues replaced with an environment-sensitive fluorescent label reveal the stabilization of the i-motif structure near the i-motif-duplex junction. These results highlight the importance of the natural environment of i-motifs for the correct assessment of their stability.
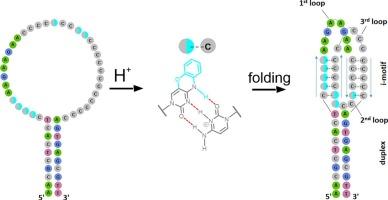
i-motif 在双链中折叠的动力学。
非典型核酸结构具有选择性地与蛋白质相互作用的能力,从而对各种细胞内过程产生影响。大量研究表明,基因组 G-四链体和 i-位点参与了转录的调控。这些结构是在 DNA 双螺旋解旋过程中临时形成的,直接测定它们是一项相当困难的任务。此外,i-motif 的折叠与 pH 值有关,大多数 i-motif 在中性 pH 值下稳定性较低。然而,一些具有长胞嘧啶重复序列的基因组 i-motif 在 pH 值为 7.3 时是稳定的,这表明它们在细胞核内具有功能性。在这里,我们研究了一个带有侧翼序列的模型 i-motif的pH依赖性行为,它形成了一个双链图案。对胞嘧啶残基被环境敏感性荧光标签取代的双模结构进行的动力学研究表明,i-motif 结构在 i-motif 双链交界处附近趋于稳定。这些结果凸显了 i-motifs 的自然环境对于正确评估其稳定性的重要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Biophysical chemistry
生物-生化与分子生物学
CiteScore
6.10
自引率
10.50%
发文量
121
审稿时长
20 days
期刊介绍:
Biophysical Chemistry publishes original work and reviews in the areas of chemistry and physics directly impacting biological phenomena. Quantitative analysis of the properties of biological macromolecules, biologically active molecules, macromolecular assemblies and cell components in terms of kinetics, thermodynamics, spatio-temporal organization, NMR and X-ray structural biology, as well as single-molecule detection represent a major focus of the journal. Theoretical and computational treatments of biomacromolecular systems, macromolecular interactions, regulatory control and systems biology are also of interest to the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


