Efficient generation of protein pockets with PocketGen
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Designing protein-binding proteins is critical for drug discovery. However, artificial-intelligence-based design of such proteins is challenging due to the complexity of protein–ligand interactions, the flexibility of ligand molecules and amino acid side chains, and sequence–structure dependencies. We introduce PocketGen, a deep generative model that produces residue sequence and atomic structure of the protein regions in which ligand interactions occur. PocketGen promotes consistency between protein sequence and structure by using a graph transformer for structural encoding and a sequence refinement module based on a protein language model. The graph transformer captures interactions at multiple scales, including atom, residue and ligand levels. For sequence refinement, PocketGen integrates a structural adapter into the protein language model, ensuring that structure-based predictions align with sequence-based predictions. PocketGen can generate high-fidelity protein pockets with enhanced binding affinity and structural validity. It operates ten times faster than physics-based methods and achieves a 97% success rate, defined as the percentage of generated pockets with higher binding affinity than reference pockets. Additionally, it attains an amino acid recovery rate exceeding 63%. A generative model that leverages a graph transformer and protein language model to generate residue sequences and full-atom structures of protein pockets is introduced, which outperforms state-of-the-art approaches.
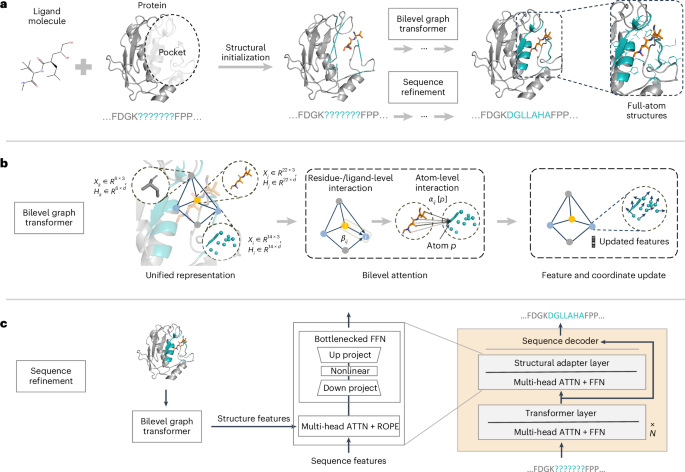
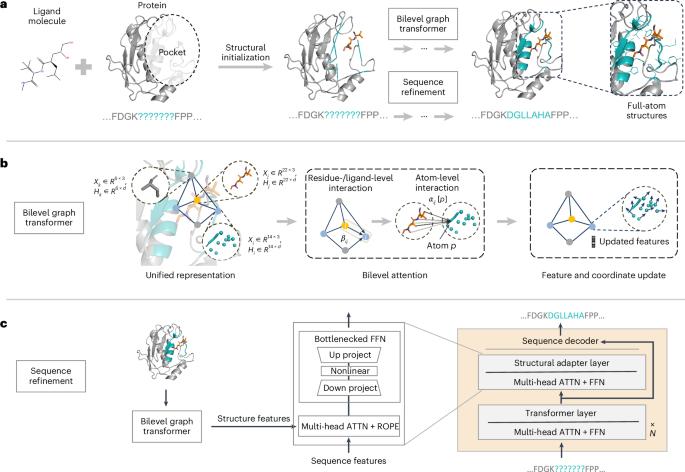
利用 PocketGen 高效生成蛋白质口袋
设计蛋白质结合蛋白对药物发现至关重要。然而,由于蛋白质-配体相互作用的复杂性、配体分子和氨基酸侧链的灵活性以及序列-结构的依赖性,基于人工智能的此类蛋白质设计极具挑战性。我们介绍的 PocketGen 是一种深度生成模型,它能生成发生配体相互作用的蛋白质区域的残基序列和原子结构。PocketGen 通过使用结构编码的图转换器和基于蛋白质语言模型的序列细化模块,促进蛋白质序列和结构之间的一致性。图转换器捕捉多个尺度的相互作用,包括原子、残基和配体水平。在序列细化方面,PocketGen 将结构适配器集成到蛋白质语言模型中,确保基于结构的预测与基于序列的预测相一致。PocketGen 可以生成高保真蛋白质口袋,增强结合亲和力和结构有效性。它的运行速度比基于物理的方法快十倍,成功率高达 97%,成功率的定义是生成的口袋的结合亲和力高于参考口袋的百分比。此外,它的氨基酸回收率超过 63%。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


