A phylogenomic tree of wood-warblers (Aves: Parulidae): Dealing with good, bad, and ugly samples
IF 3.6
1区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The New World warblers (Parulidae) are a model group for ecological and evolutionary analyses. However, current phylogenetic relationships across this family are based upon few loci. Here we use ultraconserved elements (UCEs) to estimate a rigorous species-level phylogeny for the family. As is true for many groups, high-quality tissues were unavailable for some taxa. Thus, we explored methods for incorporating sequences derived from historical (toe pad) samples to expand the phylogenetic datasets. We recovered an average of 4,186 UCE loci and mitochondrial bycatch data (supplemented with published mitochondrial data) from 96% of all currently recognized species. We found that the UCE phylogeny built with alignments with less than 70% of gaps and ambiguities recovered the most robust phylogenetic relationships for this family, representing 101 species. Using this phylogeny as a topological backbone and adding ten fair quality “bad” samples effectively generated an overall well supported phylogeny, representing 108 species (∼90% of all species). Based on this tree, we then added in seven poor quality “ugly” samples and six of those were placed within their expected genera. We also explored the phylogenetic positions of the likely extinct Leucopeza semperi and the endangered Catharopeza bishopi where limited data was obtained. Overall, taxonomic placements in our UCE trees largely correspond to previously published studies with the recovery of all currently recognized genera as monophyletic except for Basileuterus which was rendered paraphyletic by B. lachrymosus. Our study provides insights in understanding the phylogenetic relationships of a model Passeriformes family and outlines effective practices for managing sparse genomic data sourced from historical museum specimens. Variable topological arrangements across datasets and analyses reflect the evolutionary complexity of this group and provide future topics for in-depth studies.
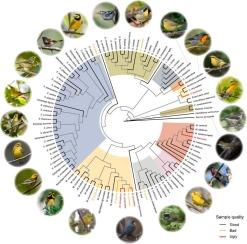
啄木鸟(鸟类:Parulidae)的系统发生树:处理好、坏、丑样本。
新世界莺科(Parulidae)是生态学和进化分析的典范类群。然而,目前该科的系统发生关系仅基于少数位点。在这里,我们使用超保守元素(UCE)来估算该科严格的物种水平系统发生。与许多类群一样,一些类群无法获得高质量的组织。因此,我们探索了从历史(趾垫)样本中获取序列的方法,以扩展系统发生学数据集。我们从目前所有已确认物种中的 96% 恢复了平均 4,186 个 UCE 位点和线粒体旁系亲属数据(辅以已发表的线粒体数据)。我们发现,用缺口和模糊性小于 70% 的排列建立的 UCE 系统发生恢复了该科最稳健的系统发生关系,代表了 101 个物种。将这一系统发生作为拓扑骨干,并添加 10 个质量一般的 "坏 "样本,有效地生成了一个总体支持良好的系统发生,代表 108 个物种(占所有物种的 90%)。在此系统树的基础上,我们又加入了 7 个质量较差的 "丑陋 "样本,其中 6 个样本被归入了预期的属。我们还探讨了可能已经灭绝的 Leucopeza semperi 和濒危的 Catharopeza bishopi 的系统发育位置,因为我们获得的数据有限。总体而言,我们的 UCE 树中的分类位置与之前发表的研究结果基本一致,除了 Basileuterus 被 B. lachrymosus 变成旁系外,所有目前公认的属都恢复了单系。我们的研究为了解一个典型的雀形目家族的系统发生关系提供了深入的见解,并为管理从博物馆历史标本中获取的稀少基因组数据提供了有效的方法。不同数据集和分析中的拓扑结构变化反映了该类群进化的复杂性,并为今后的深入研究提供了课题。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
7.50
自引率
7.30%
发文量
249
审稿时长
7.5 months
期刊介绍:
Molecular Phylogenetics and Evolution is dedicated to bringing Darwin''s dream within grasp - to "have fairly true genealogical trees of each great kingdom of Nature." The journal provides a forum for molecular studies that advance our understanding of phylogeny and evolution, further the development of phylogenetically more accurate taxonomic classifications, and ultimately bring a unified classification for all the ramifying lines of life. Phylogeographic studies will be considered for publication if they offer EXCEPTIONAL theoretical or empirical advances.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


