Autoencoder-based drug synergy framework for malignant diseases
IF 2.6
4区 生物学
Q2 BIOLOGY
引用次数: 0
Abstract
Drug combination emerges as a viable option for the treatment of malignant diseases. Drug combination outperforms monotherapy by improving therapeutic efficacy, reducing toxicity, and overcoming drug resistance. To find viable drug combinations it is difficult to traverse empirically because of enormous combinational space. Machine learning and deep learning approaches are used to uncover novel synergistic drug combinations in enormous combinational space. Here, AESyn, a novel autoencoder-based drug synergy framework for malignant diseases using a bag of words encoding is proposed. The bag of word encoding technique is used to extract drug-targeted genes. The framework utilized screening data from NCI-ALMANAC, and O’Neil datasets. Autoencoders take drug embeddings with drug-targeted genes as input for processing. The autoencoder in the proposed framework is used to extract drug features. The proposed framework is evaluated on classification and regression metrics. The performance of the proposed framework is compared with existing methods of drug synergy. According to the findings, the proposed framework achieved high performance with an accuracy of 95%, AUROC of 94.2%, and MAPE of 7.2. The autoencoder-based framework for malignant diseases using an encoding technique provides a stable, order-independent drug synergy prediction.
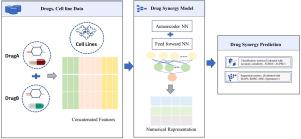
基于自动编码器的恶性疾病药物协同作用框架。
联合用药已成为治疗恶性疾病的可行方案。通过提高疗效、降低毒性和克服耐药性,联合用药优于单一疗法。由于存在巨大的组合空间,要找到可行的药物组合很难通过经验进行追踪。机器学习和深度学习方法可用于在巨大的组合空间中发现新型协同药物组合。在此,我们提出了一种基于自动编码器的新型药物协同框架 AESyn,该框架采用词袋编码技术,适用于恶性疾病。词袋编码技术用于提取药物靶向基因。该框架利用了 NCI-ALMANAC 和 O'Neil 数据集的筛选数据。自动编码器将带有药物靶向基因的药物嵌入作为输入进行处理。拟议框架中的自动编码器用于提取药物特征。拟议框架在分类和回归指标上进行了评估。将拟议框架的性能与现有的药物协同方法进行了比较。结果表明,拟议框架的准确率高达 95%,AUROC 为 94.2%,MAPE 为 7.2。基于自动编码器的恶性疾病框架使用编码技术提供了一种稳定的、与阶次无关的药物协同作用预测方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


