Multi-task learning with cross-task consistency for improved depth estimation in colonoscopy
IF 10.7
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
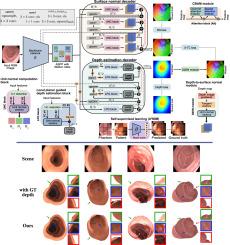
Colonoscopy screening is the gold standard procedure for assessing abnormalities in the colon and rectum, such as ulcers and cancerous polyps. Measuring the abnormal mucosal area and its 3D reconstruction can help quantify the surveyed area and objectively evaluate disease burden. However, due to the complex topology of these organs and variable physical conditions, for example, lighting, large homogeneous texture, and image modality estimating distance from the camera (aka depth) is highly challenging. Moreover, most colonoscopic video acquisition is monocular, making the depth estimation a non-trivial problem. While methods in computer vision for depth estimation have been proposed and advanced on natural scene datasets, the efficacy of these techniques has not been widely quantified on colonoscopy datasets. As the colonic mucosa has several low-texture regions that are not well pronounced, learning representations from an auxiliary task can improve salient feature extraction, allowing estimation of accurate camera depths. In this work, we propose to develop a novel multi-task learning (MTL) approach with a shared encoder and two decoders, namely a surface normal decoder and a depth estimator decoder. Our depth estimator incorporates attention mechanisms to enhance global context awareness. We leverage the surface normal prediction to improve geometric feature extraction. Also, we apply a cross-task consistency loss among the two geometrically related tasks, surface normal and camera depth. We demonstrate an improvement of 15.75% on relative error and 10.7% improvement on accuracy over the most accurate baseline state-of-the-art Big-to-Small (BTS) approach. All experiments are conducted on a recently released C3VD dataset, and thus, we provide a first benchmark of state-of-the-art methods on this dataset.
具有跨任务一致性的多任务学习,用于改进结肠镜检查的深度估计。
结肠镜检查是评估结肠和直肠异常(如溃疡和癌息肉)的黄金标准程序。测量异常粘膜区域及其三维重建有助于量化检查区域并客观评估疾病负担。然而,由于这些器官的拓扑结构复杂,物理条件多变,例如光照、大面积均匀纹理和图像模式,估计与摄像机的距离(又称深度)极具挑战性。此外,大多数结肠镜视频采集都是单眼的,这使得深度估计成为一个棘手的问题。虽然在自然场景数据集上已经提出并推进了计算机视觉深度估算方法,但这些技术在结肠镜数据集上的功效尚未得到广泛量化。由于结肠粘膜有几个纹理不明显的低纹理区域,从辅助任务中学习表征可以改善突出特征提取,从而准确估计摄像头深度。在这项工作中,我们提议开发一种新颖的多任务学习(MTL)方法,该方法具有一个共享编码器和两个解码器,即表面法线解码器和深度估计解码器。我们的深度估计器结合了注意力机制,以增强全局上下文意识。我们利用表面法线预测来改进几何特征提取。此外,我们还在两个与几何相关的任务(表面法线和摄像头深度)之间应用了跨任务一致性损失。我们证明,与最精确的最先进的大到小(BTS)方法相比,相对误差提高了 15.75%,δ1.25 精确度提高了 10.7%。所有实验都是在最近发布的 C3VD 数据集上进行的,因此,我们首次在该数据集上提供了最先进方法的基准。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


