The impact of PTEN status on glioblastoma multiforme: A glial cell type-specific study identifies unique prognostic markers
IF 7
2区 医学
Q1 BIOLOGY
引用次数: 0
Abstract
Glioblastoma multiforme (GBM) is the most invasive form of brain tumor, accounting for 5 % of the cases per 100,000 people in various countries. The phosphatase and tensin homolog deleted from chromosome 10 (PTEN) is a well-known tumor suppressor, and its alteration leads to a deleterious effect on GBM progression. The molecular mechanism of tumorigenesis in glial cell types, driven by PTEN status, is yet to be elucidated. In this study, we analyzed publicly available single-cell transcriptome profiles of PTEN wild-type (WT) and NULL GBM patients. We compared them with normal brain data to uncover many unique gene sets influenced by PTEN status. The co-expression network analysis of differentially expressed genes (DEGs) between normal brain and PTEN (WT and NULL) identified highly interconnected genes. The weighted gene co-expression network analysis (WGCNA), based on the DESeq2 algorithm, identified glial cell-type-specific modules in PTEN status-dependent bulk RNA expression profiles. We overlapped network module gene sets from single-cell and bulk transcriptome profiles, and shared genes were considered for further analysis. The hallmark pathway enrichment analysis of the genes unique to PTEN-WT and NULL revealed various tumor growth-related pathways across the glial cell types. Further characterization of PTEN-WT and PTEN-NULL networks belonging to the single-cell and bulk RNA datasets revealed that PTEN status influences the network modules in astrocytes, microglia, and oligodendrocyte precursor cells. An integrated influence value algorithm identified hub genes for each glial cell type. The prognostic analysis identified clinically relevant hub genes specific to the cell type in PTEN-WT: GLIPR2 (astrocytes), CFH, IL32, MXRA5 (microglia), and PTEN-NULL: ID1 (astrocytes) and LAT2 (microglia). Our glial cell type-level transcriptome analysis unearthed unique molecular pathways and prognostic markers in PTEN status-dependent GBM patients.
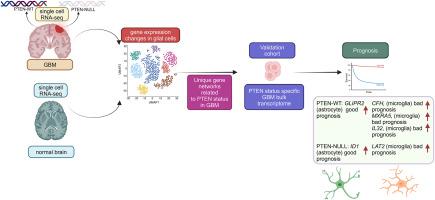
PTEN 状态对多形性胶质母细胞瘤的影响:胶质细胞类型特异性研究确定了独特的预后标志物。
多形性胶质母细胞瘤(GBM)是侵袭性最强的脑肿瘤,在各国每 10 万人中占 5%。从 10 号染色体上删除的磷酸酶和天丝同源物(PTEN)是一种众所周知的肿瘤抑制因子,它的改变会对 GBM 的进展产生有害影响。PTEN状态驱动胶质细胞类型肿瘤发生的分子机制尚待阐明。在本研究中,我们分析了可公开获得的 PTEN 野生型(WT)和 NULL GBM 患者的单细胞转录组图谱。我们将它们与正常大脑数据进行了比较,发现了许多受 PTEN 状态影响的独特基因组。对正常大脑与 PTEN(WT 和 NULL)之间差异表达基因(DEGs)的共表达网络分析发现了高度相互关联的基因。基于 DESeq2 算法的加权基因共表达网络分析(WGCNA)在 PTEN 状态依赖的大量 RNA 表达谱中发现了胶质细胞类型特异性模块。我们将单细胞和大量转录组图谱中的网络模块基因组重叠,并考虑对共享基因进行进一步分析。对PTEN-WT和NULL特有基因的标志性通路富集分析显示了神经胶质细胞类型中与肿瘤生长相关的各种通路。对属于单细胞和大量RNA数据集的PTEN-WT和PTEN-NULL网络的进一步表征显示,PTEN状态影响了星形胶质细胞、小胶质细胞和少突胶质细胞前体细胞的网络模块。综合影响值算法确定了每种神经胶质细胞类型的枢纽基因。预后分析确定了 PTEN-WT 细胞类型特有的临床相关中心基因:GLIPR2(星形胶质细胞)、CFH、IL32、MXRA5(小胶质细胞),以及 PTEN-NULL:ID1(星形胶质细胞)和 LAT2(小胶质细胞)。我们的神经胶质细胞类型水平转录组分析发现了 PTEN 状态依赖型 GBM 患者的独特分子通路和预后标志物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


