Molecular characterization of human HSPCs with different cell fates in vivo using single-cell transcriptome analysis and lentiviral barcoding technology
Abstract
Hematopoietic stem and progenitor cells (HSPCs) possess the potential to produce all types of blood cells throughout their lives. It is well recognized that HSPCs are heterogeneous, which is of great significance for their clinical applications and the treatment of diseases associated with HSPCs. This study presents a novel technology called Single-Cell transcriptome Analysis and Lentiviral Barcoding (SCALeBa) to investigate the molecular mechanisms underlying the heterogeneity of human HSPCs in vivo. The SCALeBa incorporates a transcribed barcoding library and algorithm to analyze the individual cell fates and their gene expression profiles simultaneously. Our findings using SCALeBa reveal that HSPCs subset with stronger stemness highly expressed MYL6B, ATP2A2, MYO19, MDN1, ING3, and so on. The high expression of COA3, RIF1, RAB14, and GOLGA4 may contribute to the pluripotent-lineage differentiation of HSPCs. Moreover, the roles of the representative genes revealed in this study regarding the stemness of HPSCs were confirmed with biological experiments. HSPCs expressing MRPL23 and RBM4 genes may contribute to differentiation bias into myeloid and lymphoid lineage, respectively. In addition, transcription factor (TF) characteristics of lymphoid and myeloid differentiation bias HSPCs subsets were identified and linked to previously identified genes. Furthermore, the stemness, pluripotency, and differentiation-bias genes identified with SCALeBa were verified in another independent HSPCs dataset. Finally, this study proposes using the SCALeBa-generated tracking trajectory to improve the accuracy of pseudo-time analysis results. In summary, our study provides valuable insights for understanding the heterogeneity of human HSPCs in vivo and introduces a novel technology, SCALeBa, which holds promise for broader applications.
Key points
- SCALeBa and its algorithm are developed to study the molecular mechanism underlying human HSPCs identity and function.
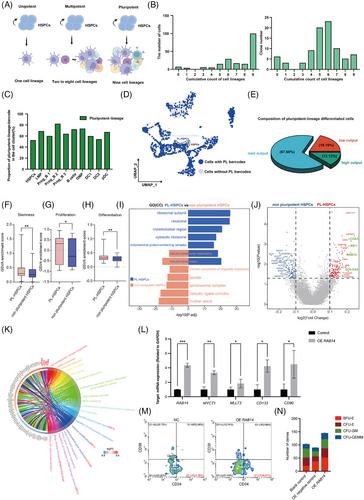
- The human HSPCs expressing MYL6B, MYO19, ATP2A2, MDN1, ING3, and PHF20 may have the capability for high stemness.
- The human HSPCs expressing COA3, RIF1, RAB14, and GOLGA4 may have the capability for pluripotent-lineage differentiation.
- The human HSPCs expressing MRPL23 and RBM4 genes may have the capability to differentiate into myeloid and lymphoid lineage respectively in vivo.
- The legitimacy of the identified genes with SCALeBa was validated using biological experiments and a public human HSPCs dataset.
- SCALeBa improves the accuracy of differentiation trajectories in monocle2-based pseudo-time analysis.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


