A mother-child data linkage approach using data from the information system for the development of research in primary care (SIDIAP) in Catalonia
IF 4
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Background
Large-scale clinical databases containing routinely collected electronic health records (EHRs) data are a valuable source of information for research studies. For example, they can be used in pharmacoepidemiology studies to evaluate the effects of maternal medication exposure on neonatal and pediatric outcomes. Yet, this type of studies is infeasible without proper mother–child linkage.
Methods
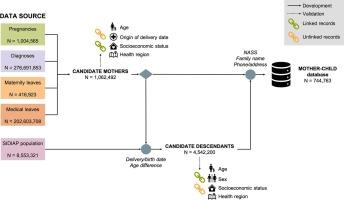
We leveraged all eligible active records (N = 8,553,321) of the Information System for Research in Primary Care (SIDIAP) database. Mothers and infants were linked using a deterministic approach and linkage accuracy was evaluated in terms of the number of records from candidate mothers that failed to link. We validated the mother–child links identified by comparison of linked and unlinked records for both candidate mothers and descendants. Differences across these two groups were evaluated by means of effect size calculations instead of p-values. Overall, we described our data linkage process following the GUidance for Information about Linking Data sets (GUILD) principles.
Results
We were able to identify 744,763 unique mother–child relationships, linking 83.8 % candidate mothers with delivery dates within a period of 15 years. Of note, we provide a record-level category label used to derive a global confidence metric for the presented linkage process. Our validation analysis showed that the two groups were similar in terms of a number of aggregated attributes.
Conclusions
Complementing the SIDIAP database with mother–child links will allow clinical researchers to expand their epidemiologic studies with the ultimate goal of improving outcomes for pregnant women and their children. Importantly, the reported information at each step of the data linkage process will contribute to the validity of analyses and interpretation of results in future studies using this resource.
利用加泰罗尼亚初级保健研究发展信息系统(SIDIAP)的数据进行母婴数据链接的方法。
背景:包含常规收集的电子健康记录(EHR)数据的大型临床数据库是研究的宝贵信息来源。例如,它们可用于药物流行病学研究,以评估母体药物暴露对新生儿和儿科预后的影响。然而,如果没有适当的母婴联系,这类研究是不可行的:我们利用了初级医疗研究信息系统(SIDIAP)数据库中所有符合条件的有效记录(N = 8,553,321)。采用确定性方法连接母亲和婴儿,并根据未能连接的候选母亲记录数量评估连接的准确性。我们通过比较候选母亲和后代的链接记录和未链接记录,验证了所识别的母婴链接。我们通过计算效应大小而不是 p 值来评估这两组之间的差异。总之,我们按照《数据集链接信息指南》(GUILD)的原则描述了我们的数据链接过程:我们能够识别 744 763 个独特的母子关系,将 83.8% 的候选母亲与 15 年内的分娩日期联系起来。值得注意的是,我们提供了一个记录级别的类别标签,用于为提出的链接过程推导出一个全局置信度指标。我们的验证分析表明,两组数据在一些综合属性方面具有相似性:通过母婴链接对 SIDIAP 数据库进行补充,将使临床研究人员能够扩大流行病学研究,最终改善孕妇及其子女的预后。重要的是,在数据链接过程的每一步所报告的信息都将有助于提高分析的有效性,并有助于今后使用这一资源进行研究时对结果的解释。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


