In vivo models of subclonal oncogenesis and dependency in hematopoietic malignancy
IF 48.8
1区 医学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Cancer evolution is a multifaceted process leading to dysregulation of cellular expansion and differentiation through somatic mutations and epigenetic dysfunction. Clonal expansion and evolution is driven by cell-intrinsic and -extrinsic selective pressures, which can be captured with increasing resolution by single-cell and bulk DNA sequencing. Despite the extensive genomic alterations revealed in profiling studies, there remain limited experimental systems to model and perturb evolutionary processes. Here, we integrate multi-recombinase tools for reversible, sequential mutagenesis from premalignancy to leukemia. We demonstrate that inducible Flt3 mutations differentially cooperate with Dnmt3a, Idh2, and Npm1 mutant alleles, and that changing the order of mutations influences cellular and transcriptional landscapes. We next use a generalizable, reversible approach to demonstrate that mutation reversion results in rapid leukemic regression with distinct differentiation patterns depending upon co-occurring mutations. These studies provide a path to experimentally model sequential mutagenesis, investigate mechanisms of transformation and probe oncogenic dependency in disease evolution.
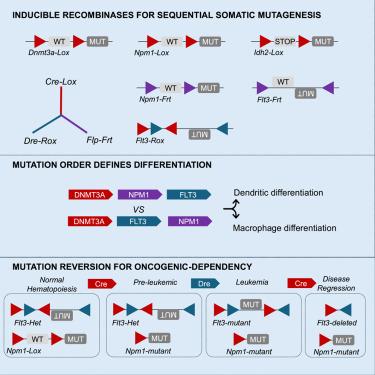
造血恶性肿瘤亚克隆肿瘤发生和依赖性的体内模型
癌症进化是一个多方面的过程,通过体细胞突变和表观遗传功能障碍导致细胞扩增和分化失调。克隆扩增和进化是由细胞内在和外在的选择性压力驱动的,单细胞和大块DNA测序可以越来越高的分辨率捕捉到这些压力。尽管分析研究揭示了广泛的基因组变化,但用于模拟和扰动进化过程的实验系统仍然有限。在这里,我们整合了多种重组酶工具,用于从恶性肿瘤前期到白血病的可逆、连续诱变。我们证明,可诱导的 Flt3 突变与 Dnmt3a、Idh2 和 Npm1 突变等位基因有不同的合作关系,改变突变的顺序会影响细胞和转录景观。接下来,我们使用一种可推广、可逆的方法证明,突变逆转会导致白血病的快速消退,而不同的分化模式取决于同时出现的突变。这些研究为实验模拟顺序突变、研究转化机制和探究疾病进化中的致癌依赖性提供了一条途径。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cancer Cell
医学-肿瘤学
CiteScore
55.20
自引率
1.20%
发文量
179
审稿时长
4-8 weeks
期刊介绍:
Cancer Cell is a journal that focuses on promoting major advances in cancer research and oncology. The primary criteria for considering manuscripts are as follows:
Major advances: Manuscripts should provide significant advancements in answering important questions related to naturally occurring cancers.
Translational research: The journal welcomes translational research, which involves the application of basic scientific findings to human health and clinical practice.
Clinical investigations: Cancer Cell is interested in publishing clinical investigations that contribute to establishing new paradigms in the treatment, diagnosis, or prevention of cancers.
Insights into cancer biology: The journal values clinical investigations that provide important insights into cancer biology beyond what has been revealed by preclinical studies.
Mechanism-based proof-of-principle studies: Cancer Cell encourages the publication of mechanism-based proof-of-principle clinical studies, which demonstrate the feasibility of a specific therapeutic approach or diagnostic test.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


