Rapid growth of acquired UBA1 mutations predisposes male patients to low-risk MDS
IF 12.8
1区 医学
Q1 HEMATOLOGY
引用次数: 0

获得性 UBA1 突变的快速增长使男性患者易患低风险 MDS
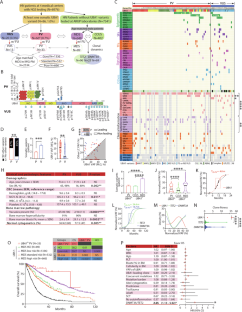
2022 年 6 月至 2023 年 11 月期间,美国四家医疗中心对血液或骨髓样本进行了有针对性的下一代测序(NGS)。我们在 86 名患者(1%,图 1A)中发现了 27 个不同的推测体细胞 UBA1 变异。66名患者(0.7%)携带九种不同的致病/可能致病变异体(PV,图 1B 蛋白序列上方)。大多数是典型的起始密码子缺失变异:p.M41T(24 例)、p.M41L(21 例)和 p.M41V(14 例),其次是之前报道的 p.M41 密码子上游的 c.118-1 G>C, 2 例)和两个新的剪接位点变异(c.118-10_118-1 del 和 c.118-5_118-1 del)。此外,三个导致 VEXAS 的错义变异 p.Y55H(N = 1)、p.G477A(N = 1)和 p.A478S(N = 1)也被归类为 PV [1、2、4]。其余 20 名患者(图 1B、C,患者 67-86)中还发现了 18 个不同的新型变异(图 1B 和补充表 1 中蛋白质序列下方),被归类为意义不确定的变异(VUS),其中包括两个复发性变异(p.D506N 和 p.I890F)。31 名 UBA1 PV 患者(47%)至少出现了一种伴随变异,与 VUS 患者(85%,p = 0.04,图 1C、D 和表 1)相比频率明显较低,同时体细胞突变负荷也较低,体细胞突变负荷定义为每名患者的体细胞变异数(图 1E,平均值 ± SEM,PV 患者为 2.0 ± 0.2,VUS 患者为 4.0 ± 0.5,p = 0.0001)。PV患者的UBA1克隆大小(图1F,平均值±SEM,26.1%±1.5)明显大于VUS患者(16.0%±3.3,p = 0.002)。55 例 PV 患者(83%)的 UBA1 变体 VAF 值高于主要并发变体(如果有的话)的 VAF 值,这表明 UBA1 PV 是创始克隆。相比之下,只有 40% 的 VUS(图 1G,p = 0.001)是主导克隆。在 PV 患者中,DNMT3A 是最常见的突变基因(23%),其次是 TET2(12%)和 ASXL1(6%,图 1C)。在 VUS 患者中,最常见的伴随变异是 TET2(补充图 1A,35%,p = 0.03),其次是 ASXL1(25%,p = 0.02)和 DNMT3A 变异(10%,p = 0.03)。值得注意的是,涉及酪氨酸激酶或RAS信号通路的变异在VUS患者中的发生率明显更高(补充图1A-C)。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


