DSDPFlex: Flexible-Receptor Docking with GPU Acceleration
IF 5.6
2区 化学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
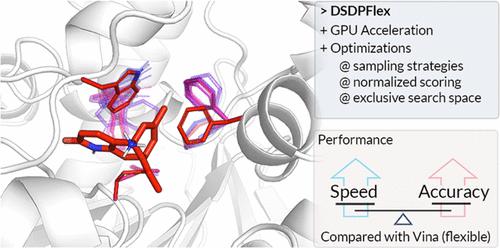
Molecular docking is an essential tool in structure-based drug discovery, widely utilized to model ligand–protein interactions and enrich potential hits. Among the different docking strategies, semiflexible docking (rigid-receptor and flexible-ligand model) is the most popular, benefiting from its balance of docking accuracy and speed. However, this approach ignores the conformational changes of proteins and hence demands suitable protein conformations as input. When the binding interaction adheres to an induced-fit model, flexible methods such as molecular dynamics simulation can be utilized, but they are computationally demanding. To balance between speed and accuracy, the flexible docking approach is an effective choice, as exemplified by AutoDock Vina and AutoDockFR, which treat selected protein side chains as flexible parts. However, the efficiency of flexible docking methods is yet to be improved for virtual screening usage. In this article, we introduce DSDPFlex, an improved flexible-receptor docking method accelerated by GPU parallelization. Beyond acceleration, optimizations with respect to sampling, scoring, and search space are implemented in DSDPFlex to further improve its capability in flexible tasks. In cross-docking evaluation, DSDPFlex demonstrates superior accuracy compared to AutoDock Vina and is 100 times faster than Vina in flexible-receptor tasks. We also show the advantage of flexible-receptor methods on suboptimal pockets and validate the advantage of DSDPFlex in screening on apo and AlphaFold2-predicted structures. With improvements in both efficiency and accuracy, DSDPFlex is expected to hold potential in future docking-based studies.
DSDPFlex:利用 GPU 加速的灵活受体对接
分子对接是基于结构的药物发现的重要工具,被广泛用于模拟配体与蛋白质之间的相互作用,并富集潜在的新药。在不同的对接策略中,半柔性对接(刚性-受体和柔性-配体模型)最受欢迎,因为它兼顾了对接的准确性和速度。然而,这种方法忽略了蛋白质的构象变化,因此需要合适的蛋白质构象作为输入。当结合相互作用遵循诱导拟合模型时,可以利用分子动力学模拟等灵活的方法,但这些方法对计算要求较高。为了在速度和准确性之间取得平衡,灵活的对接方法是一种有效的选择,AutoDock Vina 和 AutoDockFR 就是很好的例子,它们将选定的蛋白质侧链作为灵活的部分。然而,在虚拟筛选中,灵活对接方法的效率还有待提高。在本文中,我们将介绍一种通过 GPU 并行化加速的改进型柔性受体对接方法 DSDPFlex。除了加速之外,DSDPFlex 还对采样、评分和搜索空间进行了优化,以进一步提高其在灵活任务中的能力。在交叉对接评估中,与 AutoDock Vina 相比,DSDPFlex 表现出更高的准确性,在灵活的受体任务中比 Vina 快 100 倍。我们还展示了灵活受体方法在次优口袋方面的优势,并验证了 DSDPFlex 在筛选 apo 和 AlphaFold2 预测结构方面的优势。随着效率和准确性的提高,DSDPFlex有望在未来基于对接的研究中大显身手。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.80
自引率
10.70%
发文量
529
审稿时长
1.4 months
期刊介绍:
The Journal of Chemical Information and Modeling publishes papers reporting new methodology and/or important applications in the fields of chemical informatics and molecular modeling. Specific topics include the representation and computer-based searching of chemical databases, molecular modeling, computer-aided molecular design of new materials, catalysts, or ligands, development of new computational methods or efficient algorithms for chemical software, and biopharmaceutical chemistry including analyses of biological activity and other issues related to drug discovery.
Astute chemists, computer scientists, and information specialists look to this monthly’s insightful research studies, programming innovations, and software reviews to keep current with advances in this integral, multidisciplinary field.
As a subscriber you’ll stay abreast of database search systems, use of graph theory in chemical problems, substructure search systems, pattern recognition and clustering, analysis of chemical and physical data, molecular modeling, graphics and natural language interfaces, bibliometric and citation analysis, and synthesis design and reactions databases.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


