Determinants in the HTLV-1 Capsid Major Homology Region that are Critical for Virus Particle Assembly
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
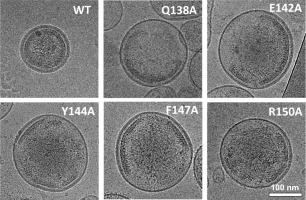
The Gag protein of retroviruses is the primary driver of virus particle assembly. Particle morphologies among retroviral genera are distinct, with intriguing differences observed relative to human immunodeficiency virus type 1 (HIV-1), particularly that of human T-cell leukemia virus type 1 (HTLV-1). In contrast to HIV-1 and other retroviruses where the capsid (CA) carboxy-terminal domain (CTD) possesses the key amino acid determinants involved in driving Gag-Gag interactions, we have previously demonstrated that the amino-terminal domain (NTD) encodes the key residues crucial for Gag multimerization and immature particle production. Here in this study, we sought to thoroughly interrogate the conserved HTLV-1 major homology region (MHR) of the CACTD to determine whether this region harbors residues important for particle assembly. In particular, site-directed mutagenesis of the HTLV-1 MHR was conducted, and mutants were analyzed for their ability to impact Gag subcellular distribution, particle production and morphology, as well as the CA-CA assembly kinetics. Several key residues (i.e., Q138, E142, Y144, F147 and R150), were found to significantly impact Gag multimerization and particle assembly. Taken together, these observations imply that while the HTLV-1 CANTD acts as the major region involved in CA-CA interactions, residues in the MHR can impact Gag multimerization, particle assembly and morphology, and likely play an important role in the conformation the CACTD that is required for CA-CA interactions.
对病毒粒子组装至关重要的 HTLV-1 荚膜主要同源区的决定因素。
逆转录病毒的 Gag 蛋白是病毒粒子组装的主要驱动力。逆转录病毒属之间的粒子形态各不相同,与 HIV-1 相比,尤其是人类 T 细胞白血病病毒 1 型(HTLV-1)的粒子形态存在着令人费解的差异。HIV-1 和其他逆转录病毒的噬菌体(CA)羧基末端结构域(CTD)具有参与驱动 Gag-Gag 相互作用的关键氨基酸决定因子,与此不同的是,我们以前已经证明氨基末端结构域(NTD)编码了对 Gag 多聚化和未成熟颗粒生成至关重要的关键残基。在本研究中,我们试图彻底检查 CACTD 的保守 HTLV-1 主要同源区 (MHR),以确定该区域是否含有对粒子组装至关重要的残基。特别是,我们对 HTLV-1 MHR 进行了定点突变,并分析了突变体对 Gag 亚细胞分布、颗粒生成和形态以及 CA-CA 组装动力学的影响。研究发现,几个关键残基(即 Q138、E142、Y144、F147 和 R150)对 Gag 的多聚化和颗粒组装有显著影响。综上所述,这些观察结果表明,虽然 HTLV-1 CANTD 是参与 CA-CA 相互作用的主要区域,但 MHR 中的残基也会影响 Gag 的多聚化、粒子组装和形态,并可能在 CA-CA 相互作用所需的 CACTD 构象中发挥重要作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


