Genome-first determination of the prevalence and penetrance of eight germline myeloid malignancy predisposition genes: a study of two population-based cohorts
IF 12.8
1区 医学
Q1 HEMATOLOGY
引用次数: 0
Abstract
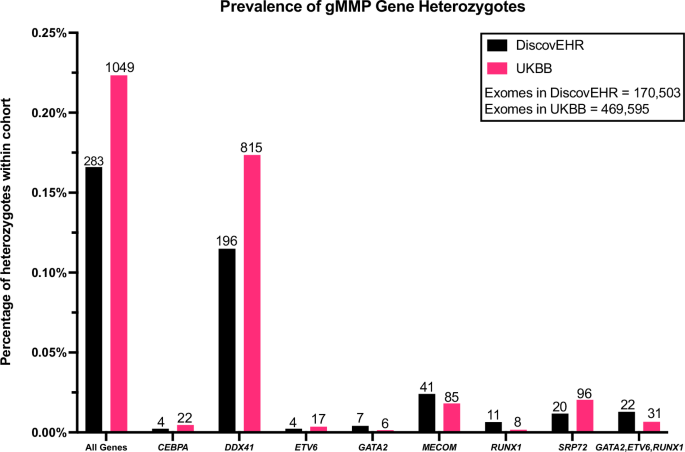
It is estimated that 10% of individuals with a myeloid malignancy carry a germline susceptibility. Using the genome-first approach, in which individuals were ascertained on genotype alone, rather than clinical phenotype, we quantified the prevalence and penetrance of pathogenic germline variants in eight myeloid malignancy predisposition (gMMP) genes. ANKRD26, CEBPA, DDX41, MECOM, SRP72, ETV6, RUNX1 and GATA2, were analyzed from the Geisinger MyCode DiscovEHR (n = 170,503) and the United Kingdom Biobank (UKBB, n = 469,595). We identified a high risk of myeloid malignancies (MM) (odds ratio[OR] all genes: DiscovEHR, 4.6 [95% confidential interval (CI) 2.1–9.7], p < 0.0001; UKBB, 6.0 [95% CI 4.3–8.2], p = 3.1 × 10-27), and decreased overall survival (hazard ratio [HR] DiscovEHR, 1.8 [95% CI 1.3–2.6], p = 0.00049; UKBB, 1.4 [95% CI 1.2–1.8], p = 8.4 × 10-5) amongst heterozygotes. Pathogenic DDX41 variants were the most commonly identified, and in UKBB showed a significantly increased risk of MM (OR 5.7 [95% CI 3.9–8.3], p = 6.0 × 10-20) and increased all-cause mortality (HR 1.35 [95% CI 1.1–1.7], p = 0.0063). Through a genome-first approach, this study genetically ascertained individuals with a gMMP and determined their MM risk and survival.

通过基因组首次确定八个种系髓系恶性肿瘤易感基因的流行率和渗透率:对两个人群队列的研究。
据估计,10% 的髓系恶性肿瘤患者携带种系易感性。采用基因组优先方法,即仅根据基因型而非临床表型确定个体,我们量化了八个髓系恶性肿瘤易感基因(gMMP)中致病性种系变异的流行率和渗透率。我们对 Geisinger MyCode DiscovEHR(n = 170,503)和英国生物库(UKBB,n = 469,595)中的 ANKRD26、CEBPA、DDX41、MECOM、SRP72、ETV6、RUNX1 和 GATA2 进行了分析。我们发现罹患骨髓恶性肿瘤(MM)的风险很高(所有基因的几率比[OR]:DiscovEHR,4.6 [95% 保密区间 (CI) 2.1-9.7],p -27),杂合子的总生存率降低(危险比 [HR] DiscovEHR,1.8 [95% CI 1.3-2.6],p = 0.00049;UKBB,1.4 [95% CI 1.2-1.8],p = 8.4 × 10-5)。致病性 DDX41 变体是最常见的变体,在 UKBB 中显示 MM 风险显著增加(OR 5.7 [95% CI 3.9-8.3],p = 6.0 × 10-20),全因死亡率增加(HR 1.35 [95% CI 1.1-1.7],p = 0.0063)。本研究通过基因组优先的方法,从基因上确定了具有 gMMP 的个体,并确定了他们的 MM 风险和存活率。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


