CACHE Challenge #1: Targeting the WDR Domain of LRRK2, A Parkinson’s Disease Associated Protein
IF 5.6
2区 化学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
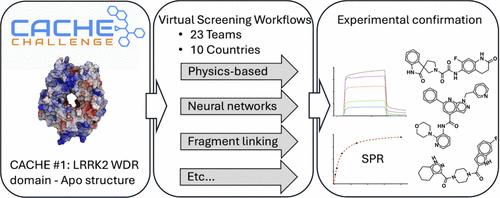
The CACHE challenges are a series of prospective benchmarking exercises to evaluate progress in the field of computational hit-finding. Here we report the results of the inaugural CACHE challenge in which 23 computational teams each selected up to 100 commercially available compounds that they predicted would bind to the WDR domain of the Parkinson’s disease target LRRK2, a domain with no known ligand and only an apo structure in the PDB. The lack of known binding data and presumably low druggability of the target is a challenge to computational hit finding methods. Of the 1955 molecules predicted by participants in Round 1 of the challenge, 73 were found to bind to LRRK2 in an SPR assay with a KD lower than 150 μM. These 73 molecules were advanced to the Round 2 hit expansion phase, where computational teams each selected up to 50 analogs. Binding was observed in two orthogonal assays for seven chemically diverse series, with affinities ranging from 18 to 140 μM. The seven successful computational workflows varied in their screening strategies and techniques. Three used molecular dynamics to produce a conformational ensemble of the targeted site, three included a fragment docking step, three implemented a generative design strategy and five used one or more deep learning steps. CACHE #1 reflects a highly exploratory phase in computational drug design where participants adopted strikingly diverging screening strategies. Machine learning-accelerated methods achieved similar results to brute force (e.g., exhaustive) docking. First-in-class, experimentally confirmed compounds were rare and weakly potent, indicating that recent advances are not sufficient to effectively address challenging targets.
CACHE 挑战 #1:靶向帕金森病相关蛋白 LRRK2 的 WDR 结构域
CACHE 挑战赛是一系列前瞻性的基准测试活动,旨在评估计算寻找靶点领域的进展。在这里,我们报告了首届 CACHE 挑战赛的结果,23 个计算团队各自选择了多达 100 个市售化合物,预测这些化合物将与帕金森病靶标 LRRK2 的 WDR 结构域结合。该结构域没有已知配体,在 PDB 中只有 apo 结构。由于缺乏已知的结合数据,该靶点的可药性可能很低,这对计算寻找靶点的方法是一个挑战。在挑战赛第一轮参与者预测的 1955 个分子中,有 73 个分子在 SPR 试验中与 LRRK2 结合,KD 低于 150 μM。这 73 个分子进入了第 2 轮的 "命中扩展 "阶段,每个计算小组最多选择 50 个类似物。在两个正交试验中观察到了七个化学性质不同的系列的结合,亲和力从 18 μM 到 140 μM。七个成功的计算工作流程在筛选策略和技术方面各不相同。三个使用分子动力学方法生成了目标位点的构象组合,三个包含了片段对接步骤,三个实施了生成设计策略,五个使用了一个或多个深度学习步骤。CACHE #1反映了计算药物设计的高度探索阶段,参与者采用了截然不同的筛选策略。机器学习加速方法取得了与蛮力(如穷举)对接相似的结果。经实验证实的第一类化合物非常罕见,且药效较弱,这表明最近的进步还不足以有效解决具有挑战性的靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.80
自引率
10.70%
发文量
529
审稿时长
1.4 months
期刊介绍:
The Journal of Chemical Information and Modeling publishes papers reporting new methodology and/or important applications in the fields of chemical informatics and molecular modeling. Specific topics include the representation and computer-based searching of chemical databases, molecular modeling, computer-aided molecular design of new materials, catalysts, or ligands, development of new computational methods or efficient algorithms for chemical software, and biopharmaceutical chemistry including analyses of biological activity and other issues related to drug discovery.
Astute chemists, computer scientists, and information specialists look to this monthly’s insightful research studies, programming innovations, and software reviews to keep current with advances in this integral, multidisciplinary field.
As a subscriber you’ll stay abreast of database search systems, use of graph theory in chemical problems, substructure search systems, pattern recognition and clustering, analysis of chemical and physical data, molecular modeling, graphics and natural language interfaces, bibliometric and citation analysis, and synthesis design and reactions databases.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


