Computational study of the HLTF ATPase remodeling domain suggests its activity on dsDNA and implications in damage tolerance
IF 2.7
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The Helicase-Like Transcription Factor (HLTF) is member of the SWI/SNF-family of ATP dependent chromatin remodellers known primarily for maintaining genome stability. Biochemical and cellular assays support its multiple roles in DNA Damage Tolerance. However, the lack of sufficient structural data limits the comprehension of the molecular basis of its modes of action.
In this work we have modelled and characterized the HLTF ATPase remodeling domain by using bioinformatic tools and all-atoms molecular dynamics simulations.
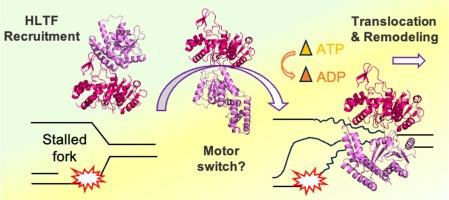
In-silico results suggested that its binding to dsDNA is mainly mediated by the positively charged residues Arg563 and Lys913, found conserved in HLTF homologs, and Arg620 and Lys999, found only in HLTF. Interestingly, these residues are mutated in cancer cells. During translocation on dsDNA, HLTF remains persistently bound through the N-terminal ATPase subunit. However, DNA advancement occurs only in the presence of the synergic-anticorrelated action of both motor lobes. In contrast, the C-terminal facilitates substrate remodeling through DNA deformation and generation of bulges according to a wave-model. Finally, the large conformational change suggested between the two motor-remodeling subunits might be activated upon the release of PARP1 on stalled fork and be responsible for the intervention of HLTF-HIRAN in the formation of D-loop and 4-way junction DNA structures.
对 HLTF ATPase 重塑结构域的计算研究表明了它在 dsDNA 上的活性以及对损伤耐受性的影响。
类螺旋酶转录因子(HLTF)是 ATP 依赖性染色质重塑因子 SWI/SNF 家族的成员,主要用于维持基因组的稳定性。生化和细胞测定支持其在 DNA 损伤耐受中的多重作用。然而,由于缺乏足够的结构数据,限制了对其作用模式的分子基础的理解。在这项工作中,我们利用生物信息学工具和全原子分子动力学模拟对 HLTF ATPase 重塑结构域进行了建模和表征。模拟结果表明,它与dsDNA的结合主要是由带正电荷的残基Arg563和Lys913以及Arg620和Lys999介导的,Arg563和Lys913在HLTF同源物中是保守的,而Arg620和Lys999只在HLTF中发现。有趣的是,这些残基在癌细胞中发生了突变。在dsDNA的转位过程中,HLTF通过N端ATPase亚基保持持续结合。然而,只有在两个运动叶协同作用的情况下,DNA 才会向前推进。与此相反,根据波浪模型,C 端通过 DNA 变形和产生隆起促进底物重塑。最后,两个马达重塑亚基之间的巨大构象变化可能会在停滞叉上的 PARP1 释放时被激活,并导致 HLTF-HIRAN 介入 D 环和 4 向连接 DNA 结构的形成。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of structural biology
生物-生化与分子生物学
CiteScore
6.30
自引率
3.30%
发文量
88
审稿时长
65 days
期刊介绍:
Journal of Structural Biology (JSB) has an open access mirror journal, the Journal of Structural Biology: X (JSBX), sharing the same aims and scope, editorial team, submission system and rigorous peer review. Since both journals share the same editorial system, you may submit your manuscript via either journal homepage. You will be prompted during submission (and revision) to choose in which to publish your article. The editors and reviewers are not aware of the choice you made until the article has been published online. JSB and JSBX publish papers dealing with the structural analysis of living material at every level of organization by all methods that lead to an understanding of biological function in terms of molecular and supermolecular structure.
Techniques covered include:
• Light microscopy including confocal microscopy
• All types of electron microscopy
• X-ray diffraction
• Nuclear magnetic resonance
• Scanning force microscopy, scanning probe microscopy, and tunneling microscopy
• Digital image processing
• Computational insights into structure
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


