Conformational Differences in the Light Chain Constant Domain of Immunoglobulin G and Free Light Chain May Influence Proteolysis in AL Amyloidosis
IF 4.7
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
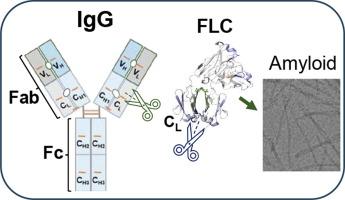
Immunoglobulin light chain amyloidosis (AL) is a life-threatening disease caused by the deposition of light chain (LC) and its fragments containing variable (VL) and portions of constant (CL) domains. AL patients feature either monoclonal free LCs (FLCs) circulating as covalent and noncovalent homodimers, or monoclonal immunoglobulin (Ig) wherein the LC and heavy chain (HC) form disulfide-linked heterodimers, or both. The role of full-length Ig in AL amyloidosis is unclear as prior studies focused on FLC or VL domain. We used a mammalian cell-based expression system to generate four AL patient-derived full-length IgGs, two non-AL IgG controls, and six corresponding FLC proteins derived from an IGLV6-57 germline precursor. Comparison of proteins’ secondary structure, thermal stability, proteolytic susceptibility, and disulfide link reduction suggested the importance of local vs. global conformational stability. Analysis of IgGs vs. corresponding FLCs using hydrogen–deuterium exchange mass spectrometry revealed major differences in the local conformation/dynamics of the CL domain. In all IgGs vs. FLCs, segments containing β-strand and α-helix βAC-αACBC were more dynamic/exposed while segment βDC-βEC was less dynamic/exposed. Notably, these segments overlapped proteolysis-prone regions whose in vivo cleavage generates LC fragments found in AL deposits. Altogether, the results suggest that preferential cleavage in segments βAC-αACBC of FLC or βDC-βEC of LC in IgG helps generate amyloid protein precursors. We propose that protecting these segments using small-molecule stabilizers, which bind to the interfacial cavities CL-CL in FLC and/or CL-CH1 in IgG, is a potential therapeutic strategy to complement current approaches targeting VL-VL or VL-CL stabilization of LC dimer.
免疫球蛋白 G 的轻链恒定域和游离轻链的构象差异可能会影响 AL 淀粉样变性病的蛋白水解。
免疫球蛋白轻链淀粉样变性(AL)是一种威胁生命的疾病,由轻链(LC)及其包含可变(VL)和部分恒定(CL)结构域的片段沉积引起。AL 患者的特征是单克隆游离轻链(FLC)以共价和非共价同二聚体形式循环,或单克隆免疫球蛋白(Ig),其中轻链和重链(HC)形成二硫键连接的异二聚体,或两者兼而有之。全长 Ig 在 AL 淀粉样变性中的作用尚不清楚,因为之前的研究主要集中在 FLC 或 VL 结构域。我们使用基于哺乳动物细胞的表达系统生成了四种 AL 患者来源的全长 IgG、两种非 AL IgG 对照以及六种相应的 FLC 蛋白,这些蛋白来源于 IGLV6-57 胚系前体。蛋白质二级结构、热稳定性、蛋白水解敏感性和二硫键还原性的比较表明了局部构象稳定性与整体构象稳定性的重要性。利用氢氘交换质谱分析 IgG 与相应的 FLC,发现 CL 结构域的局部构象/动力学存在重大差异。在所有 IgG 与 FLCs 的对比中,含有 β-链和α-螺旋 βAC-αACBC 的区段的动态/暴露程度较高,而 βDC-βEC 区段的动态/暴露程度较低。值得注意的是,这些片段与蛋白水解易发区重叠,蛋白水解易发区在体内裂解产生 AL 沉积物中的 LC 片段。总之,这些结果表明,IgG 中 FLC 的 βAC-αACBC 段或 LC 的 βDC-βEC 段的优先裂解有助于产生淀粉样蛋白前体。我们建议使用小分子稳定剂来保护这些区段,这种稳定剂可与 FLC 中的 CL-CL 和/或 IgG 中的 CL-CH1 接口空腔结合,是一种潜在的治疗策略,可补充目前以 VL-VL 或 VL-CL 稳定 LC 二聚体为目标的方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


