Molecular dynamics simulations, essential dynamics and MMPBSA to evaluate natural compounds as potential inhibitors for AccD6, a key drug target in the fatty acid biosynthesis pathway in Mycobacterium tuberculosis
IF 2.7
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
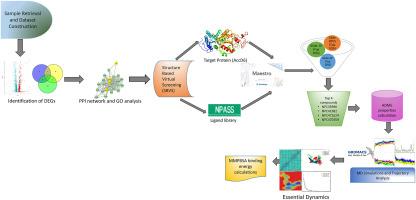
Antibiotic resistance in Mycobacterium tuberculosis, the primary causative agent of the tuberculosis disease is an ever growing threat especially in developing and underdeveloped countries. Isoniazid is a commonly used first line anti-tuberculosis drug used during the first phase of tuberculosis treatment. However, due to its improper use, many strains of Mycobacterium tuberculosis have acquired resistance to the drug. Advancements in next generation sequencing technologies, such as transcriptomics have paved way for identifying alternative drug targets based on the differential expression pattern of genes. Therefore, this study makes use of RNA-Seq data of Mycobacterium tuberculosis isolates treated with different concentrations of isoniazid to identify genes that can be proposed as drug targets. From the differential expression analysis, it was observed that four genes were significantly upregulated under all the conditions. Among the four genes, accD6 was selected as the drug target for virtual screening and molecular dynamics studies, because of its role in fatty acid elongation and contribution to the synthesis of mycolic acids. The protein-protein interaction network and gene ontology based functional enrichment studies show an enrichment in fatty acid biosynthesis related pathways. Furthermore, virtual screening studies successfully screened the top three natural inhibitor molecules with satisfactory ADME properties and a better glide score than the reference compound, NCI-172033. The trajectory analysis, essential dynamics studies and MMPBSA analysis, concluded that among the hit molecules, NPC41982, a thiazole derivative showed the most promising results and can be considered as a potential drug candidate.
利用分子动力学模拟、基本动力学和 MMPBSA 评估天然化合物作为 AccD6(结核分枝杆菌脂肪酸生物合成途径中的一个关键药物靶点)潜在抑制剂的效果
结核分枝杆菌是结核病的主要致病菌,其抗生素耐药性是一个日益严重的威胁,尤其是在发展中国家和欠发达国家。异烟肼是结核病治疗第一阶段常用的一线抗结核药物。然而,由于使用不当,许多结核分枝杆菌菌株对这种药物产生了抗药性。下一代测序技术(如转录组学)的进步为根据基因的差异表达模式确定替代药物靶点铺平了道路。因此,本研究利用用不同浓度异烟肼处理的结核分枝杆菌分离株的 RNA-Seq 数据来确定可作为药物靶点的基因。通过差异表达分析发现,有四个基因在所有条件下都显著上调。在这四个基因中,accD6因其在脂肪酸伸长过程中的作用和对霉酚酸合成的贡献而被选为虚拟筛选和分子动力学研究的药物靶标。基于蛋白质-蛋白质相互作用网络和基因本体论的功能富集研究表明,脂肪酸生物合成相关通路中存在富集。此外,虚拟筛选研究成功地筛选出了前三名天然抑制剂分子,它们具有令人满意的 ADME 特性,且滑翔得分优于参考化合物 NCI-172033。通过轨迹分析、基本动力学研究和 MMPBSA 分析,得出结论认为,在命中的分子中,噻唑衍生物 NPC41982 的结果最有希望,可作为潜在的候选药物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


