Vesicle Picker: A tool for efficient identification of membrane protein complexes in vesicles
IF 2.7
3区 生物学
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
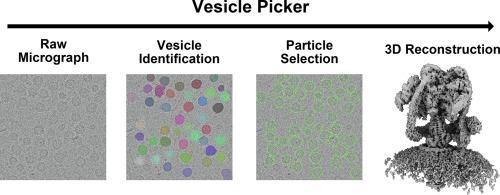
Electron cryomicroscopy (cryo-EM) has recently allowed determination of near-atomic resolution structures of membrane proteins and protein complexes embedded in lipid vesicles. However, particle selection from electron micrographs of these vesicles can be challenging due to the strong signal contributed from the lipid bilayer. This challenge often requires iterative and laborious particle selection workflows to generate a dataset of high-quality particle images for subsequent analysis. Here we present Vesicle Picker, an open-source program built on the Segment Anything model. Vesicle Picker enables automatic identification of vesicles in cryo-EM micrographs with high recall and precision. It then exhaustively selects all potential particle locations, either at the perimeter or uniformly over the surface of the projection of the vesicle. The program is designed to interface with cryoSPARC, which performs both upstream micrograph processing and downstream single particle image analysis. We demonstrate Vesicle Picker’s utility by determining a high-resolution map of the vacuolar-type ATPase from micrographs of native synaptic vesicles (SVs) and identifying an additional protein or protein complex in the SV membrane.
囊泡拾取器:有效识别囊泡中膜蛋白复合物的工具。
电子低温显微镜(cryo-EM)近来可以测定嵌入脂质囊泡中的膜蛋白和蛋白复合物的近原子分辨率结构。然而,由于脂质双分子层会产生强烈的信号,因此从这些囊泡的电子显微图像中选择颗粒是一项挑战。这一挑战往往需要反复、费力的粒子选择工作流程,以生成高质量的粒子图像数据集,用于后续分析。在此,我们介绍基于 Segment Anything 模型的开源程序 Vesicle Picker。Vesicle Picker 能自动识别低温电子显微镜显微照片中的囊泡,识别率和精确度都很高。然后,该程序会在囊泡投影的周边或均匀表面详尽选择所有潜在的颗粒位置。该程序旨在与 cryoSPARC 相连接,后者可进行上游显微图像处理和下游单颗粒图像分析。我们从原生突触囊泡 (SV) 的显微照片中确定了空泡型 ATP 酶的高分辨率图,并确定了 SV 膜中的另一种蛋白质或蛋白质复合物,从而证明了 Vesicle Picker 的实用性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of structural biology
生物-生化与分子生物学
CiteScore
6.30
自引率
3.30%
发文量
88
审稿时长
65 days
期刊介绍:
Journal of Structural Biology (JSB) has an open access mirror journal, the Journal of Structural Biology: X (JSBX), sharing the same aims and scope, editorial team, submission system and rigorous peer review. Since both journals share the same editorial system, you may submit your manuscript via either journal homepage. You will be prompted during submission (and revision) to choose in which to publish your article. The editors and reviewers are not aware of the choice you made until the article has been published online. JSB and JSBX publish papers dealing with the structural analysis of living material at every level of organization by all methods that lead to an understanding of biological function in terms of molecular and supermolecular structure.
Techniques covered include:
• Light microscopy including confocal microscopy
• All types of electron microscopy
• X-ray diffraction
• Nuclear magnetic resonance
• Scanning force microscopy, scanning probe microscopy, and tunneling microscopy
• Digital image processing
• Computational insights into structure
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


