The dynamic evolution of lineage switch under CD19 CAR-T treatment in non-KMT2A rearranged B-ALL patients
IF 12.8
1区 医学
Q1 HEMATOLOGY
引用次数: 0

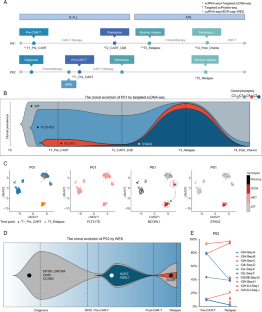
非 KMT2A 重排 B-ALL 患者在 CD19 CAR-T 治疗下的系谱转换动态演变
重建克隆进化范式有助于我们理解世系转换的过程[6]。因此,我们通过单细胞靶向DNA测序描绘了患者1(P01)的克隆进化模式(补充表3)。通过单细胞基因组测序和质量控制,我们在四个时间点共获得了 6566 个具有相关基因突变(FLT3、BCORL1 和 STAG2)的高质量细胞,用于克隆结构推断(图 1B,补充表 4)。Tapestri insights 分析(图 1B)显示,FLT3-ITD 突变在四个不同的时间点持续存在(83.3%、69.4%、98.8%、9.0%)。在骨髓复发开始后,原有的 BCORL1 突变迅速扩大,而 STAG2 突变与 BCORL1 突变同时出现(16.1%、4.3%、96.1%、0.0%)。与 T1_Pre_CART 时间点相比,我们发现 BCORL1 突变负荷(图 1C)在 T3_Relapse 时间点有所增加。此外,单细胞膜蛋白数据(补充图1A-D,补充表5)显示,在T1_Pre_CART时间点,B-ALL爆破细胞表达典型的B-ALL相关标志物CD19和CD10,并同时表达髓系相关标志物CD33和CD123,而在复发时,爆破细胞失去了淋巴标志物CD19的表达,证实了系的转换现象。此外,通过全外显子测序和靶向测序重建了患者2(P02)的克隆进化结构(图1D,补充表6)。在P02的整个治疗过程中,融合基因EP300::ZNF384持续表达,IKZF2突变克隆在CD19 CAR-T治疗后消失,BCOR基因突变克隆在髓系复发后出现。通过 BCR 测序,我们观察到在 T2_Relapse 时间点存在与 T1_Pre_CART 时间点相同的免疫球蛋白序列(图 1E,补充表 7),而且我们还观察到在复发过程中免疫球蛋白序列的克隆频率增加,这表明它们在髓系重编程过程中富集。这一结果表明,髓系祖细胞的起源是从 B-ALL 细胞重编程而来[7]。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Leukemia
医学-血液学
CiteScore
18.10
自引率
3.50%
发文量
270
审稿时长
3-6 weeks
期刊介绍:
Title: Leukemia
Journal Overview:
Publishes high-quality, peer-reviewed research
Covers all aspects of research and treatment of leukemia and allied diseases
Includes studies of normal hemopoiesis due to comparative relevance
Topics of Interest:
Oncogenes
Growth factors
Stem cells
Leukemia genomics
Cell cycle
Signal transduction
Molecular targets for therapy
And more
Content Types:
Original research articles
Reviews
Letters
Correspondence
Comments elaborating on significant advances and covering topical issues
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


