HEART: Learning better representation of EHR data with a heterogeneous relation-aware transformer
IF 4
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
Objective:
Pretrained language models have recently demonstrated their effectiveness in modeling Electronic Health Record (EHR) data by modeling the encounters of patients as sentences. However, existing methods fall short of utilizing the inherent heterogeneous correlations between medical entities—which include diagnoses, medications, procedures, and lab tests. Existing studies either focus merely on diagnosis entities or encode different entities in a homogeneous space, leading to suboptimal performance. Motivated by this, we aim to develop a foundational language model pre-trained on EHR data with explicitly incorporating the heterogeneous correlations among these entities.
Methods:
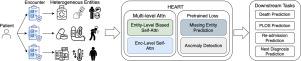
In this study, we propose HEART, a heterogeneous relation-aware transformer for EHR. Our model includes a range of heterogeneous entities within each input sequence and represents pairwise relationships between entities as a relation embedding. Such a higher-order representation allows the model to perform complex reasoning and derive attention weights in the heterogeneous context. Additionally, a multi-level attention scheme is employed to exploit the connection between different encounters while alleviating the high computational costs. For pretraining, HEART engages with two tasks, missing entity prediction and anomaly detection, which both effectively enhance the model’s performance on various downstream tasks.
Results:
Extensive experiments on two EHR datasets and five downstream tasks demonstrate HEART’s superior performance compared to four SOTA foundation models. For instance, HEART achieves improvements of 12.1% and 4.1% over Med-BERT in death and readmission prediction, respectively. Additionally, case studies show that HEART offers interpretable insights into the relationships between entities through the learned relation embeddings.
Conclusion:
We study the problem of EHR representation learning and propose HEART, a model that leverages the heterogeneous relationships between medical entities. Our approach includes a multi-level encoding scheme and two specialized pretrained objectives, designed to boost both the efficiency and effectiveness of the model. We have comprehensively evaluated HEART across five clinically significant downstream tasks using two EHR datasets. The experimental results verify the model’s great performance and validate its practical utility in healthcare applications. Code: https://github.com/Graph-and-Geometric-Learning/HEART.
HEART:利用异构关系感知转换器学习更好的电子病历数据表示。
目的:最近,预训练语言模型通过将患者的就诊情况建模为句子,证明了其在电子健康记录(EHR)数据建模方面的有效性。然而,现有的方法无法利用医疗实体(包括诊断、药物、手术和化验)之间固有的异质性关联。现有研究要么只关注诊断实体,要么在同质空间中对不同实体进行编码,从而导致性能不佳。受此启发,我们旨在开发一种在电子病历数据上进行预训练的基础语言模型,明确纳入这些实体之间的异质相关性:在这项研究中,我们提出了 HEART,一种用于电子病历的异构关系感知转换器。我们的模型包括每个输入序列中的一系列异构实体,并将实体间的成对关系表示为关系嵌入。这种高阶表示法允许模型在异构环境中执行复杂的推理并推导出关注权重。此外,HEART 还采用了多级注意力方案,以利用不同遭遇之间的联系,同时降低高昂的计算成本。在预训练中,HEART 参与了缺失实体预测和异常检测两项任务,这两项任务都能有效提高模型在各种下游任务中的性能:在两个电子病历数据集和五个下游任务上进行的广泛实验表明,与四个 SOTA 基础模型相比,HEART 的性能更为出色。例如,在死亡预测和再入院预测方面,HEART 比 Med-BERT 分别提高了 12.1% 和 4.1%。此外,案例研究表明,HEART 通过学习到的关系嵌入对实体之间的关系提供了可解释的见解:我们对电子病历表示学习问题进行了研究,并提出了 HEART 模型,该模型充分利用了医疗实体之间的异构关系。我们的方法包括多级编码方案和两个专门的预训练目标,旨在提高模型的效率和有效性。我们利用两个电子病历数据集对 HEART 的五项临床重要下游任务进行了全面评估。实验结果验证了该模型的卓越性能,并验证了其在医疗保健应用中的实用性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


