The potential clinical utility of Whole Genome Sequencing for patients with cancer: evaluation of a regional implementation of the 100,000 Genomes Project
IF 6.4
1区 医学
Q1 ONCOLOGY
引用次数: 0
Abstract
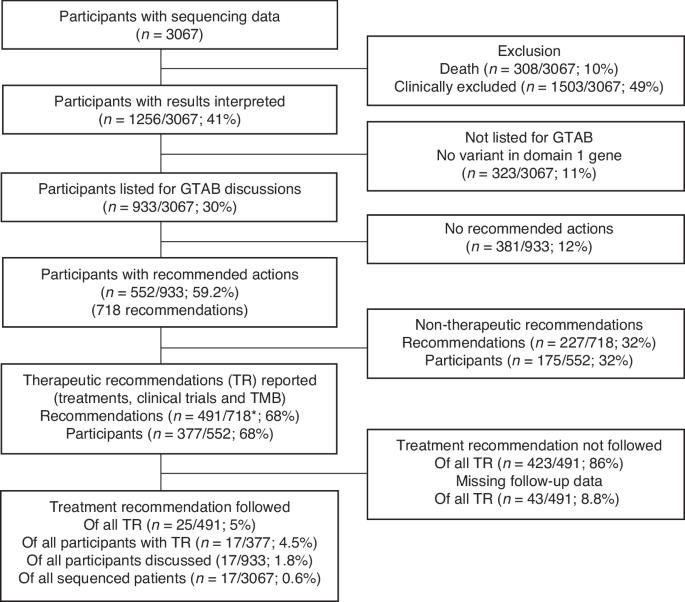
The 100,000 Genomes Project established infrastructure for Whole Genome Sequencing (WGS) in the United Kingdom. A retrospective study of cancer patients recruited to the 100,000 Genomes Project by the West Midlands Genomics Medicine Centre, evaluating clinical relevance of results. After excluding samples with no sequencing data (1678/4851; 34.6%), 3166 sample sets (germline and somatic) from 3067 participants were sequenced. Results of 1256 participants (41.0%) were interpreted (excluding participants who died (308/3067; 10.0%) or were clinically excluded (1503/3067; 49.0%)). Of these, 323 (25.7%) had no variants in genes which may alter management (Domain 1 genes). Of the remaining 933 participants, 552 (59.2%) had clinical recommendations made (718 recommendations in total). These included therapeutic recommendations (377/933; 40.4%), such as clinical trial, unlicensed or licensed therapies or high TMB recommendations, and germline variants warranting clinical genetics review (85/933; 9.1%). At the last follow up, 20.2% of all recommendations were followed (145/718). However, only a small proportion of therapeutic recommendations were followed (5.1%, 25/491). The 100,000 Genomes Project has established infrastructure and regional experience to support personalised cancer care. The majority of those with successful sequencing had actionable variants. Ensuring GTAB recommendations are followed will maximise benefits for patients.
全基因组测序对癌症患者的潜在临床效用:对十万基因组计划地区实施情况的评估。
背景:10 万基因组计划在英国建立了全基因组测序(WGS)基础设施:十万基因组计划在英国建立了全基因组测序(WGS)基础设施:对西米德兰兹基因组医学中心(West Midlands Genomics Medicine Centre)纳入十万基因组计划的癌症患者进行回顾性研究,评估结果的临床相关性:在排除没有测序数据的样本(1678/4851;34.6%)后,对来自3067名参与者的3166个样本集(种系和体细胞)进行了测序。对 1256 名参与者(41.0%)的结果进行了解读(不包括死亡参与者(308/3067;10.0%)或临床排除参与者(1503/3067;49.0%))。其中,323 人(25.7%)的基因中没有可能改变管理的变异(领域 1 基因)。在其余 933 名参与者中,有 552 人(59.2%)获得了临床建议(共 718 项建议)。其中包括治疗建议(377/933;40.4%),如临床试验、无证或有证疗法或高 TMB 建议,以及需要进行临床遗传学审查的种系变异(85/933;9.1%)。在最后一次随访中,20.2%的建议得到了遵从(145/718)。然而,只有一小部分治疗建议得到了遵循(5.1%,25/491):结论:十万基因组计划建立了支持个性化癌症治疗的基础设施和地区经验。大多数成功测序者都有可操作的变异。确保遵循GTAB的建议将为患者带来最大益处。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

British Journal of Cancer
医学-肿瘤学
CiteScore
15.10
自引率
1.10%
发文量
383
审稿时长
6 months
期刊介绍:
The British Journal of Cancer is one of the most-cited general cancer journals, publishing significant advances in translational and clinical cancer research.It also publishes high-quality reviews and thought-provoking comment on all aspects of cancer prevention,diagnosis and treatment.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


