Branched-chain α-ketoacids aerobically activate HIF1α signalling in vascular cells
IF 18.9
1区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
Abstract
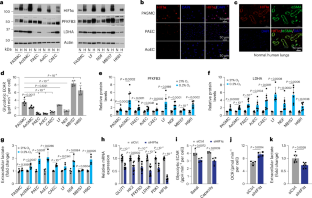
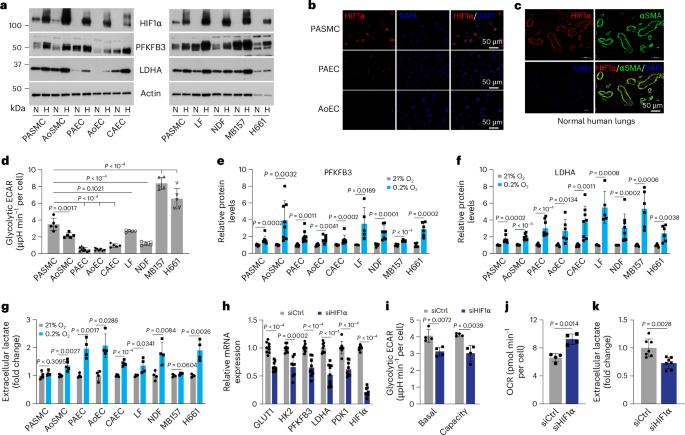
Hypoxia-inducible factor 1α (HIF1α) is a master regulator of biological processes in hypoxia. Yet, the mechanisms and biological consequences of aerobic HIF1α activation by intrinsic factors, particularly in normal (primary) cells, remain elusive. Here we show that HIF1α signalling is activated in several human primary vascular cells in normoxia and in vascular smooth muscle cells of normal human lungs. Mechanistically, aerobic HIF1α activation is mediated by paracrine secretion of three branched-chain α-ketoacids (BCKAs), which suppress PHD2 activity via direct inhibition and via LDHA-mediated generation of L-2-hydroxyglutarate. BCKA-mediated HIF1α signalling activation stimulated glycolytic activity and governed a phenotypic switch of pulmonary artery smooth muscle cells, which correlated with BCKA metabolic dysregulation and pathophenotypic changes in pulmonary arterial hypertension patients and male rat models. We thus identify BCKAs as previously unrecognized signalling metabolites that aerobically activate HIF1α and that the BCKA–HIF1α pathway modulates vascular smooth muscle cell function, an effect that may be relevant to pulmonary vascular pathobiology. Branched-chain α-ketoacids are shown to aerobically activate HIF1α signalling, which induces a phenotypic switch in vascular smooth muscle cells that is potentially relevant in the context of pulmonary artery hypertension.
支链α-酮酸有氧激活血管细胞中的 HIF1α 信号传导
缺氧诱导因子 1α(HIF1α)是缺氧状态下生物过程的主要调节因子。然而,内在因子激活有氧 HIF1α 的机制和生物学后果,尤其是在正常(原代)细胞中,仍然难以捉摸。在这里,我们发现在正常缺氧状态下,HIF1α 信号在几种人类原代血管细胞和正常人类肺部血管平滑肌细胞中被激活。从机理上讲,有氧 HIF1α 激活是由三种支链 α-酮酸(BCKA)的旁分泌介导的,它们通过直接抑制和 LDHA 介导的 L-2- 羟基戊二酸的生成来抑制 PHD2 的活性。BCKA 介导的 HIF1α 信号激活可刺激糖酵解活性并支配肺动脉平滑肌细胞的表型转换,这与肺动脉高压患者和雄性大鼠模型中 BCKA 代谢失调和病理表型变化相关。因此,我们发现 BCKA 是以前未曾认识到的信号代谢物,它能有氧激活 HIF1α,而且 BCKA-HIF1α 通路能调节血管平滑肌细胞的功能,这种效应可能与肺血管病理生物学有关。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature metabolism
ENDOCRINOLOGY & METABOLISM-
CiteScore
27.50
自引率
2.40%
发文量
170
期刊介绍:
Nature Metabolism is a peer-reviewed scientific journal that covers a broad range of topics in metabolism research. It aims to advance the understanding of metabolic and homeostatic processes at a cellular and physiological level. The journal publishes research from various fields, including fundamental cell biology, basic biomedical and translational research, and integrative physiology. It focuses on how cellular metabolism affects cellular function, the physiology and homeostasis of organs and tissues, and the regulation of organismal energy homeostasis. It also investigates the molecular pathophysiology of metabolic diseases such as diabetes and obesity, as well as their treatment. Nature Metabolism follows the standards of other Nature-branded journals, with a dedicated team of professional editors, rigorous peer-review process, high standards of copy-editing and production, swift publication, and editorial independence. The journal has a high impact factor, has a certain influence in the international area, and is deeply concerned and cited by the majority of scholars.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


