Image-based 3D genomics through chromatin tracing
IF 56
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
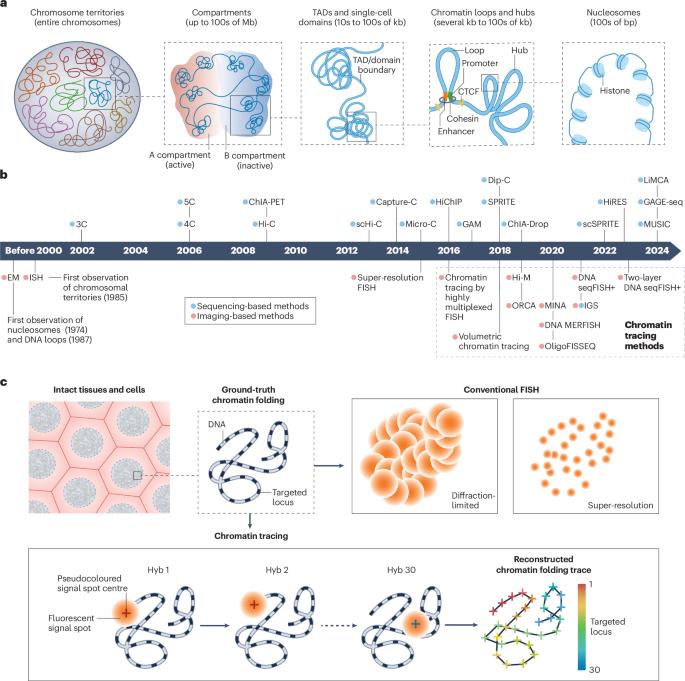
Correct organization of higher-order genome folding is essential for the regulation of gene expression, DNA replication and other genomic functions. Technological advances in high-throughput sequencing-based methods have allowed for systematic profiling of the fundamental architectural features of chromatin organization at the genome level. However, how chromatin is folded in 3D space at single-cell and single-chromosome-copy resolution in intact cells and tissues has been a long-standing question owing to a lack of appropriate methodology. Recent advances in chromatin labelling, imaging and automated fluidics technologies have led to the development of chromatin tracing, enabling direct mapping of the 3D chromatin folding trajectory in situ at the single-cell and single-molecule level. Within nearly a decade of its development, chromatin tracing has been applied at different genomic scales and to a spectrum of cell types and model organisms, improving our understanding of the structures, mechanisms and functions of chromatin organization in various biological and medical areas. In this Primer, we introduce the experimental principles, data analysis procedures and current applications of chromatin tracing. We describe how chromatin tracing can be combined with multimodal imaging and genetic screening technologies and provide a perspective on the limitations of current chromatin tracing approaches and the direction of technological developments for filling major gaps in discoveries. Chromatin tracing involves the direct mapping of 3D chromatin folding in situ, and can be applied at a number of genomic scales and to a wide range of cell types and model organisms. In this Primer, Yang & Wang discuss the design of chromatin tracing experiments, data analysis procedures and how chromatin tracing can be combined with multi-modal imaging.
通过染色质追踪进行基于图像的三维基因组学研究
高阶基因组折叠的正确组织对于调控基因表达、DNA 复制和其他基因组功能至关重要。基于高通量测序方法的技术进步使我们能够在基因组水平上系统地分析染色质组织的基本结构特征。然而,由于缺乏适当的方法,染色质在完整细胞和组织中如何以单细胞和单染色体拷贝的分辨率在三维空间中折叠一直是一个长期存在的问题。染色质标记、成像和自动流体力学技术的最新进展促进了染色质追踪技术的发展,使其能够在单细胞和单分子水平上直接绘制染色质原位折叠轨迹的三维图。在染色质追踪技术发展的近十年间,它已被应用于不同的基因组尺度、各种细胞类型和模式生物,从而提高了我们对染色质组织结构、机制和功能在各种生物和医学领域的认识。在本入门指南中,我们将介绍染色质追踪的实验原理、数据分析程序和当前应用。我们介绍了染色质描记如何与多模态成像和基因筛选技术相结合,并透视了当前染色质描记方法的局限性以及填补重大发现空白的技术发展方向。染色质追踪涉及原位三维染色质折叠的直接制图,可应用于多种基因组尺度、多种细胞类型和模式生物。在这本《入门》中,Yang & Wang 讨论了染色质追踪实验的设计、数据分析程序以及染色质追踪如何与多模式成像相结合。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


