Implications of trinodal inhibitions and drug repurposing in MAPK pathway: A putative remedy for breast cancer
IF 2.6
4区 生物学
Q2 BIOLOGY
引用次数: 0
Abstract
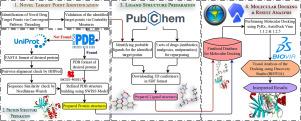
Breast cancer has been one of the supreme causes of cancer-related deaths among women worldwide. To make the case even more compounded, due to innate or acquired causes, cancer cells often develop resistance against the available chemotherapy or monotargeted treatments. This resistance is concomitant with increased activation of the MAPK (mitogen-activated protein kinase) signaling pathway. This study simultaneously targets three imperative intermediates in this pathway using molecular docking and real-time simulation. Docking was performed via the integrated AutoDock Vina 1.1.2 & 1.2.5 of the PyRx software, while the Discovery Studio (BIOVIA) v24.1.0.23298 was utilized to conduct the simulation. The aim is to investigate the therapeutic prospects of known potential inhibitors of the targeted intermediates and repurposable drugs to comprehend the effectiveness of targeting these trinodes simultaneously. The target points were deemed to be PDPK1 (3-phosphoinositide-dependent protein kinase 1), ERK1/2 (extracellular signal-related protein kinases 1/2), and mTOR (mammalian target of Rapamycin). Our study reveals that out of the candidate inhibitors chosen for each node, MP7 exhibited the most superior binding affinities for all three: −10.918 kcal/mol for PDPK1, −10.224 kcal/mol for ERK1, −10.134 kcal/mol for ERK2, and −9.2 kcal/mol for mTOR (via AutoDock Vina 1, .2.5). Some scores with MP7 were often higher than the available single-targeted drugs for different nodes in the MAPK pathway. Additionally, a total of 1867 repurposed analgesic, antibiotic, and antiparasitic drugs, including Zavegepant (−13.399 kcal/mol for PDPK1), Adozelesin (−11.74 kcal/mol for mTOR) and Modoflaner (−11.29 kcal/mol for PDPK1), showed promising binding energetics while targeting our triad points than other compounds used. This approach prompts for mitigating not only breast cancer but other elusive diseases as well, with state-of-the-art multitargeted therapies coupled with bioinformatic strategies.
MAPK 通路中的三联抑制和药物再利用的意义:乳腺癌的可能治疗方法
乳腺癌一直是全球妇女因癌症死亡的主要原因之一。更为严重的是,由于先天或后天的原因,癌细胞通常会对现有的化疗或单一靶向治疗产生抗药性。这种抗药性与 MAPK(丝裂原活化蛋白激酶)信号通路的激活增加同时发生。本研究通过分子对接和实时模拟,同时针对这一途径中的三个必要中间体进行了研究。对接是通过 PyRx 软件中集成的 AutoDock Vina 1.1.2 和 1.2.5 进行的,而模拟则是利用 Discovery Studio (BIOVIA) v24.1.0.23298 进行的。目的是研究靶向中间体的已知潜在抑制剂和可再利用药物的治疗前景,以了解同时靶向这些三节点的有效性。靶点被认为是 PDPK1(3-磷酸肌醇依赖性蛋白激酶 1)、ERK1/2(细胞外信号相关蛋白激酶 1/2)和 mTOR(雷帕霉素哺乳动物靶点)。我们的研究显示,在为每个节点选择的候选抑制剂中,MP7 对所有三种抑制剂都表现出最优越的结合亲和力:对 PDPK1 为 -10.918 kcal/mol,对 ERK1 为 -10.224 kcal/mol,对 ERK2 为 -10.134 kcal/mol,对 mTOR 为 -9.2 kcal/mol(通过 AutoDock Vina 1,.2.5)。对于 MAPK 通路的不同节点,MP7 的一些得分往往高于现有的单一靶向药物。此外,包括 Zavegepant(对 PDPK1 为 -13.399 kcal/mol)、Adozelesin(对 mTOR 为 -11.74 kcal/mol)和 Modoflaner(对 PDPK1 为 -11.29 kcal/mol)在内的总共 1867 种再利用镇痛药、抗生素和抗寄生虫药物,在靶向我们的三联体点时显示出了比其他化合物更好的结合能。这种方法不仅能缓解乳腺癌,还能通过最先进的多靶点疗法和生物信息学策略缓解其他难以治愈的疾病。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


