Improving the prediction of protein stability changes upon mutations by geometric learning and a pre-training strategy
IF 12
Q1 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
Abstract
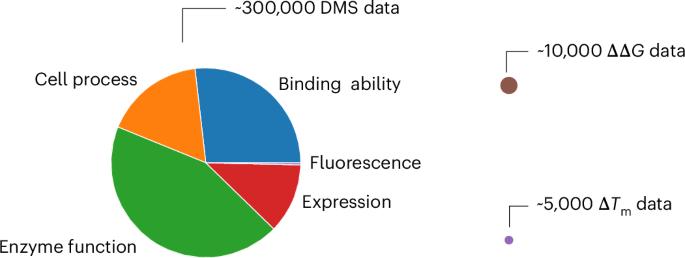
Accurate prediction of protein mutation effects is of great importance in protein engineering and design. Here we propose GeoStab-suite, a suite of three geometric learning-based models—GeoFitness, GeoDDG and GeoDTm—for the prediction of fitness score, ΔΔG and ΔTm of a protein upon mutations, respectively. GeoFitness engages a specialized loss function to allow supervised training of a unified model using the large amount of multi-labeled fitness data in the deep mutational scanning database. To further improve the downstream tasks of ΔΔG and ΔTm prediction, the encoder of GeoFitness is reutilized as a pre-trained module in GeoDDG and GeoDTm to overcome the challenge of lacking sufficient labeled data. This pre-training strategy, in combination with data expansion, markedly improves model performance and generalizability. In the benchmark test, GeoDDG and GeoDTm outperform the other state-of-the-art methods by at least 30% and 70%, respectively, in terms of the Spearman correlation coefficient. In this study, the authors propose a strategy to train a unified model to learn the general mutational effects based on multi-labeled deep mutational scanning (DMS) data, and then reutilize this pre-trained model to improve the downstream protein stability prediction tasks.
通过几何学习和预训练策略改进突变后蛋白质稳定性变化的预测。
准确预测蛋白质突变效应对蛋白质工程和设计至关重要。在此,我们提出了 GeoStab-suite,这是一套由 GeoFitness、GeoDDG 和 GeoDTm 三种基于几何学习的模型组成的套件,分别用于预测蛋白质突变后的适应度得分、ΔΔG 和ΔTm。GeoFitness 使用专门的损失函数,利用深度突变扫描数据库中的大量多标签适配性数据对统一模型进行监督训练。为了进一步改进ΔΔG和ΔTm预测的下游任务,GeoFitness的编码器被重新用作GeoDDG和GeoDTm的预训练模块,以克服缺乏足够标记数据的挑战。这种预训练策略与数据扩展相结合,显著提高了模型的性能和普适性。在基准测试中,GeoDDG 和 GeoDTm 的斯皮尔曼相关系数分别比其他先进方法高出至少 30% 和 70%。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


