Genomic and transcriptomic perspectives on the origin and evolution of NUMTs in Orthoptera
IF 3.6
1区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Nuclear mitochondrial pseudogenes (NUMTs) result from the transfer of mitochondrial DNA (mtDNA) to the nuclear genome. NUMTs, as “frozen” snapshots of mitochondria, can provide insights into diversification patterns. In this study, we analyzed the origins and insertion frequency of NUMTs using genome assembly data from ten species in Orthoptera. We found divergences between NUMTs and contemporary mtDNA in Orthoptera ranging from 0 % to 23.78 %. The results showed that the number of NUMT insertions was significantly positively correlated with the content of transposable elements in the genome. We found that 39.09 %-68.65 % of the NUMTs flanking regions (2,000 bp) contained retrotransposons, and more NUMTs originated from mitochondrial rDNA regions. Based on the analysis of the mitochondrial transcriptome, we found a potential mechanism of NUMT integration: mitochondrial transcripts are reverse transcribed into double-stranded DNA and then integrated into the genome. The probability of this mechanism occurring accounts for 0.30 %-1.02 % of total mitochondrial nuclear transfer events. Finally, based on the phylogenetic tree constructed using NUMTs and contemporary mtDNA, we provide insights into ancient evolutionary events such as species-specific “autaponumts” and “synaponumts” shared among different species, as well as post-integration duplication events.
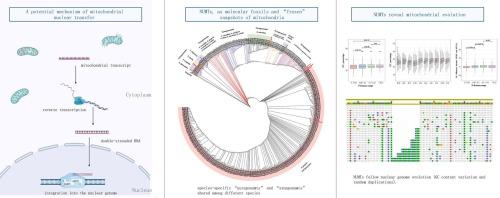
从基因组和转录组角度看直翅目 NUMTs 的起源和进化。
核线粒体假基因(NUMTs)是线粒体DNA(mtDNA)转移到核基因组的结果。NUMTs作为线粒体的 "冷冻 "快照,可以让人们深入了解线粒体的多样化模式。在这项研究中,我们利用直翅目 10 个物种的基因组组装数据分析了 NUMTs 的起源和插入频率。我们发现 NUMTs 与直翅目当代 mtDNA 之间的差异从 0 % 到 23.78 % 不等。结果表明,NUMT插入的数量与基因组中转座元件的含量呈显著正相关。我们发现,39.09%-68.65%的NUMTs侧翼区域(2000 bp)含有反转座子,更多的NUMTs来源于线粒体rDNA区域。根据对线粒体转录本组的分析,我们发现了 NUMT 整合的潜在机制:线粒体转录本反向转录为双链 DNA,然后整合到基因组中。这种机制发生的概率占线粒体核转移事件总数的 0.30%-1.02%。最后,根据利用 NUMTs 和当代 mtDNA 构建的系统发生树,我们深入了解了古老的进化事件,如不同物种之间共享的物种特异性 "自聚体 "和 "合聚体",以及整合后的复制事件。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
7.50
自引率
7.30%
发文量
249
审稿时长
7.5 months
期刊介绍:
Molecular Phylogenetics and Evolution is dedicated to bringing Darwin''s dream within grasp - to "have fairly true genealogical trees of each great kingdom of Nature." The journal provides a forum for molecular studies that advance our understanding of phylogeny and evolution, further the development of phylogenetically more accurate taxonomic classifications, and ultimately bring a unified classification for all the ramifying lines of life. Phylogeographic studies will be considered for publication if they offer EXCEPTIONAL theoretical or empirical advances.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


