TopoTxR: A topology-guided deep convolutional network for breast parenchyma learning on DCE-MRIs
IF 10.7
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Characterization of breast parenchyma in dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is a challenging task owing to the complexity of underlying tissue structures. Existing quantitative approaches, like radiomics and deep learning models, lack explicit quantification of intricate and subtle parenchymal structures, including fibroglandular tissue. To address this, we propose a novel topological approach that explicitly extracts multi-scale topological structures to better approximate breast parenchymal structures, and then incorporates these structures into a deep-learning-based prediction model via an attention mechanism. Our topology-informed deep learning model, TopoTxR, leverages topology to provide enhanced insights into tissues critical for disease pathophysiology and treatment response. We empirically validate TopoTxR using the VICTRE phantom breast dataset, showing that the topological structures extracted by our model effectively approximate the breast parenchymal structures. We further demonstrate TopoTxR’s efficacy in predicting response to neoadjuvant chemotherapy. Our qualitative and quantitative analyses suggest differential topological behavior of breast tissue in treatment-naïve imaging, in patients who respond favorably to therapy as achieving pathological complete response (pCR) versus those who do not. In a comparative analysis with several baselines on the publicly available I-SPY 1 dataset (N = 161, including 47 patients with pCR and 114 without) and the Rutgers proprietary dataset (N = 120, with 69 patients achieving pCR and 51 not), TopoTxR demonstrates a notable improvement, achieving a 2.6% increase in accuracy and a 4.6% enhancement in AUC compared to the state-of-the-art method.
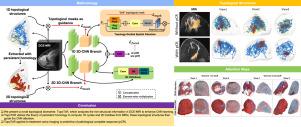
TopoTxR:拓扑引导的深度卷积网络,用于在 DCE-MRI 上学习乳腺实质。
动态对比增强磁共振成像(DCE-MRI)中乳腺实质的特征描述是一项具有挑战性的任务,因为底层组织结构非常复杂。现有的定量方法,如放射组学和深度学习模型,缺乏对包括纤维腺体组织在内的复杂而微妙的实质结构的明确量化。为了解决这个问题,我们提出了一种新颖的拓扑方法,它能明确提取多尺度拓扑结构,以更好地逼近乳腺实质结构,然后通过注意力机制将这些结构纳入基于深度学习的预测模型。我们的基于拓扑结构的深度学习模型 TopoTxR 利用拓扑结构增强了对疾病病理生理学和治疗反应关键组织的洞察力。我们使用 VICTRE 模型乳腺数据集对 TopoTxR 进行了经验验证,结果表明我们的模型提取的拓扑结构有效地接近了乳腺实质结构。我们进一步证明了 TopoTxR 在预测新辅助化疗反应方面的功效。我们的定性和定量分析结果表明,在未经治疗的成像中,乳腺组织的拓扑行为存在差异,对治疗反应良好的患者可获得病理完全反应 (pCR),而对治疗反应不佳的患者则无法获得病理完全反应 (pCR)。在对公开的 I-SPY 1 数据集(N = 161,包括 47 名获得病理完全反应的患者和 114 名未获得病理完全反应的患者)和罗格斯专有数据集(N = 120,包括 69 名获得病理完全反应的患者和 51 名未获得病理完全反应的患者)与几种基线进行的比较分析中,TopoTxR 显示出明显的改进,与最先进的方法相比,准确率提高了 2.6%,AUC 提高了 4.6%。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


