Comparative Study of Allosteric GPCR Binding Sites and Their Ligandability Potential
IF 5.6
2区 化学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
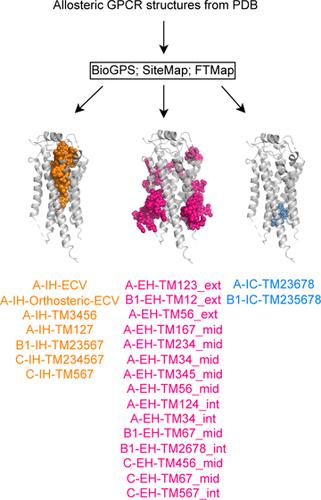
The steadily growing number of experimental G-protein-coupled receptor (GPCR) structures has revealed diverse locations of allosteric modulation, and yet few drugs target them. This gap highlights the need for a deeper understanding of allosteric modulation in GPCR drug discovery. The current work introduces a systematic annotation scheme to structurally classify GPCR binding sites based on receptor class, transmembrane helix contacts, and, for membrane-facing sites, membrane sublocation. This GPCR specific annotation scheme was applied to 107 GPCR structures bound by small molecules contributing to 24 distinct allosteric binding sites for comparative evaluation of three binding site detection methods (BioGPS, SiteMap, and FTMap). BioGPS identified the most in 22 of 24 sites. In addition, our property analysis showed that extrahelical allosteric ligands and binding sites represent a distinct chemical space characterized by shallow pockets with low volume, and the corresponding allosteric ligands showed an enrichment of halogens. Furthermore, we demonstrated that combining receptor and ligand similarity can be a viable method for ligandability assessment. One challenge regarding site prediction is the ligand shaping effect on the observed binding site, especially for extrahelical sites where the ligand-induced effect was most pronounced. To our knowledge, this is the first study presenting a binding site annotation scheme standardized for GPCRs, and it allows a comparison of allosteric binding sites across different receptors in an objective way. The insight from this study provides a framework for future GPCR binding site studies and highlights the potential of targeting allosteric sites for drug development.
异位 GPCR 结合位点及其配体潜力的比较研究
实验性 G 蛋白偶联受体(GPCR)结构的数量稳步增长,揭示了异位调节的不同位置,但针对这些位置的药物却寥寥无几。这一差距凸显了在 GPCR 药物发现过程中深入了解异调的必要性。目前的研究工作引入了一种系统注释方案,根据受体类别、跨膜螺旋接触情况以及面向膜的位点、膜亚定位对 GPCR 结合位点进行结构分类。该 GPCR 特定注释方案被应用于 107 个与小分子结合的 GPCR 结构,这些小分子构成了 24 个不同的异构结合位点,用于对三种结合位点检测方法(BioGPS、SiteMap 和 FTMap)进行比较评估。在 24 个结合位点中,BioGPS 对 22 个位点的识别率最高。此外,我们的性质分析表明,螺旋外的异生配体和结合位点代表了一个独特的化学空间,其特点是具有低容积的浅口袋,相应的异生配体显示出卤素的富集。此外,我们还证明,将受体和配体的相似性结合起来是一种可行的配体性评估方法。位点预测的一个挑战是配体对观察到的结合位点的塑造效应,特别是对于配体诱导效应最明显的螺旋外位点。据我们所知,这是第一项针对 GPCRs 提出标准化结合位点注释方案的研究,它可以客观地比较不同受体的异构结合位点。这项研究的洞察力为未来的 GPCR 结合位点研究提供了一个框架,并凸显了以异生结合位点为靶点进行药物开发的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
9.80
自引率
10.70%
发文量
529
审稿时长
1.4 months
期刊介绍:
The Journal of Chemical Information and Modeling publishes papers reporting new methodology and/or important applications in the fields of chemical informatics and molecular modeling. Specific topics include the representation and computer-based searching of chemical databases, molecular modeling, computer-aided molecular design of new materials, catalysts, or ligands, development of new computational methods or efficient algorithms for chemical software, and biopharmaceutical chemistry including analyses of biological activity and other issues related to drug discovery.
Astute chemists, computer scientists, and information specialists look to this monthly’s insightful research studies, programming innovations, and software reviews to keep current with advances in this integral, multidisciplinary field.
As a subscriber you’ll stay abreast of database search systems, use of graph theory in chemical problems, substructure search systems, pattern recognition and clustering, analysis of chemical and physical data, molecular modeling, graphics and natural language interfaces, bibliometric and citation analysis, and synthesis design and reactions databases.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


