Epitope-anchored contrastive transfer learning for paired CD8+ T cell receptor–antigen recognition
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
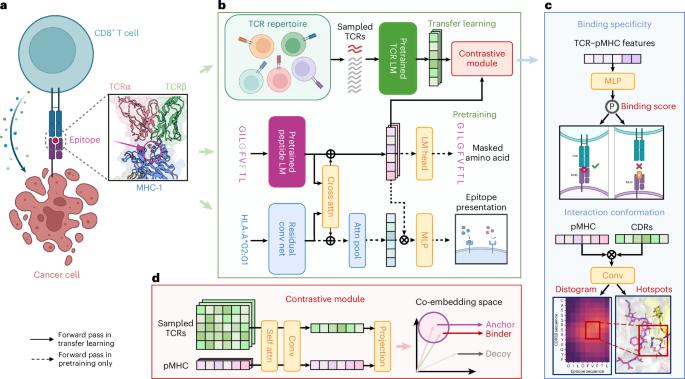
Understanding the mechanisms of T cell antigen recognition that underpin adaptive immune responses is critical for developing vaccines, immunotherapies and treatments against autoimmune diseases. Despite extensive research efforts, accurate prediction of T cell receptor (TCR)–antigen binding pairs remains a great challenge due to the vast diversity and cross-reactivity of TCRs. Here we propose a deep-learning-based framework termed epitope-anchored contrastive transfer learning (EPACT) tailored to paired human CD8+ TCRs. Harnessing the pretrained representations and co-embeddings of peptide–major histocompatibility complex (pMHC) and TCR, EPACT demonstrated generalizability in predicting binding specificity for unseen epitopes and distinct TCR repertoires. Contrastive learning enabled highly precise predictions for immunodominant epitopes and interpretable analysis of epitope-specific T cells. We applied EPACT to SARS-CoV-2-responsive T cells, and the predicted binding strength aligned well with the surge in spike-specific immune responses after vaccination. We further fine-tuned EPACT on structural data to decipher the residue-level interactions involved in TCR–antigen recognition. EPACT was capable of quantifying interchain distance matrices and identifying contact residues, corroborating the presence of TCR cross-reactivity across multiple tumour-associated antigens. Together, EPACT can serve as a useful artificial intelligence approach with important potential in practical applications and contribute towards the development of TCR-based immunotherapies. Accurate prediction of T cell receptor (TCR)–antigen recognition remains a challenge. Zhang et al. propose a contrastive transfer learning model to predict TCR–pMHC binding that enables interpretable analyses of epitope-specific T cells and can decipher residue-level interactions.
CD8+T细胞受体-抗原配对识别的表位锚定对比迁移学习
了解支撑适应性免疫反应的 T 细胞抗原识别机制对于开发疫苗、免疫疗法和治疗自身免疫性疾病至关重要。尽管开展了大量研究工作,但由于 TCR 的多样性和交叉反应性,准确预测 T 细胞受体(TCR)与抗原的结合对仍然是一项巨大的挑战。在这里,我们提出了一种基于深度学习的框架,称为表位锚定对比转移学习(EPACT),专门针对成对的人类 CD8+ TCR。利用肽-主要组织相容性复合体(pMHC)和TCR的预训练表征和共嵌入,EPACT在预测未知表位和不同TCR复合物的结合特异性方面展示了通用性。对比学习可以对免疫优势表位进行高度精确的预测,并对表位特异性 T 细胞进行可解释的分析。我们将 EPACT 应用于 SARS-CoV-2 反应性 T 细胞,预测的结合强度与接种疫苗后尖峰特异性免疫反应的激增非常吻合。我们根据结构数据进一步微调了 EPACT,以破译 TCR 与抗原识别中涉及的残基级相互作用。EPACT 能够量化链间距离矩阵并识别接触残基,从而证实多种肿瘤相关抗原之间存在 TCR 交叉反应。总之,EPACT可以作为一种有用的人工智能方法,在实际应用中具有重要潜力,并有助于开发基于TCR的免疫疗法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


